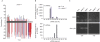RNA-directed DNA methylation and plant development require an IWR1-type transcription factor - PubMed (original) (raw)
doi: 10.1038/embor.2009.246. Epub 2009 Dec 11.
Etienne Bucher, Lucia Daxinger, Bruno Huettel, David P Kreil, Frank Breinig, Marc Lind, Manfred J Schmitt, Stacey A Simon, Sai Guna Ranjan Gurazada, Blake C Meyers, Zdravko J Lorkovic, Antonius J M Matzke, Marjori Matzke
Affiliations
- PMID: 20010803
- PMCID: PMC2816620
- DOI: 10.1038/embor.2009.246
RNA-directed DNA methylation and plant development require an IWR1-type transcription factor
Tatsuo Kanno et al. EMBO Rep. 2010 Jan.
Abstract
RNA-directed DNA methylation (RdDM) in plants requires two RNA polymerase (Pol) II-related RNA polymerases, namely Pol IV and Pol V. A genetic screen designed to reveal factors that are important for RdDM in a developmental context in Arabidopsis identified DEFECTIVE IN MERISTEM SILENCING 4 (DMS4). Unlike other mutants defective in RdDM, dms4 mutants have a pleiotropic developmental phenotype. The DMS4 protein is similar to yeast IWR1 (interacts with RNA polymerase II), a conserved putative transcription factor that interacts with Pol II subunits. The DMS4 complementary DNA partly complements the K1 killer toxin hypersensitivity of a yeast iwr1 mutant, suggesting some functional conservation. In the transgenic system studied, mutations in DMS4 directly or indirectly affect Pol IV-dependent secondary short interfering RNAs, Pol V-mediated RdDM, Pol V-dependent synthesis of intergenic non-coding RNA and expression of many Pol II-driven genes. These data suggest that DMS4 might be a regulatory factor for several RNA polymerases, thus explaining its diverse roles in the plant.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
Figure 1
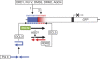
Transgene system and stepwise RdDM. The target enhancer (blue bar) and downstream region (shaded red bar) undergo stepwise RdDM. In the first step, 24 nt primary siRNAs (blue dashes), which are derived by DCL3 processing of a Pol II-generated hairpin RNA, induce primary RdDM (blue ‘m') at the target enhancer region. This requires the SNF2-like factor DRD1, Pol V, DMS3 (a structural maintenance of chromosomes hinge-domain-containing protein; Kanno et al, 2008), AGO4 (Daxinger et al, 2009) and presumably the de novo methyltransferase DRM2. In the second step, methylated DNA is directly or indirectly transcribed by Pol IV, producing an enhancer-associated ‘nascent' RNA (solid black arrow) that is turned over in a pathway involving RDR2 and DCL3 to generate 24 nt secondary siRNAs (red dashes), which trigger secondary RdDM (red ‘m') in the downstream region. Whether AGO slicing of the ‘nascent' RNA (broken black arrow) is required to produce aberrant RNA substrates for RDR2 is unknown. Primary RdDM is sufficient to silence expression of the downstream GFP reporter gene (Daxinger et al, 2009). AGO, ARGONAUTE; DCL3, DICER-LIKE 3; DMS3, DEFECTIVE IN MERISTEM SILENCING 3; DRD1, DEFECTIVE IN RNA-DIRECTED DNA METHYLATION 1; DRM2, DOMAINS REARRANGED METHYLTRANSFERASE 2; GFP, green fluorescent protein; RdDM, RNA-directed DNA methylation; RDR2, RNA-DEPENDENT RNA POLYMERASE 2; siRNA, small interfering RNA; SNF2, sucrose non-fermenting 2.
Figure 2
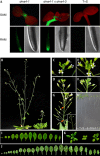
Effects of dms4 mutations on GFP silencing and plant development. (A) Mutations in the DMS4 gene release GFP silencing in shoot and root apical meristems (SAM and RAM, respectively). High fluorescence is seen in the hypocotyl at this stage of development (Kanno et al, 2008). (B) Phenotypes of adult dms4-1 mutant (right) and wild-type plants (left). Although flower morphology seems normal in the dms4 mutants, (C,D) floral inflorescences of wild-type plants are less closely packed than (E,F) inflorescences of the dms4 mutant. (G) Short internode distances and disturbed phyllotaxy (orange arrows) of the dms4 mutant; normal spiral phyllotaxy is shown inset. (H) dms4-1 seedlings (right) are pale and grow more slowly than wild-type seedlings (left), suggesting a chloroplast defect. (I) Rosette leaves of wild-type plants and (J) the dms4 mutant at the time of bolting. (K) Young plants of the wild-type (left) and dms4-1 and dms4-3 mutants (middle and right, respectively). DMS4, DEFECTIVE IN MERISTEM SILENCING 4; dms4-1, seedling homozygous for the dms4-1 allele; dms4-1 × dms4-3, heteroallelic combination; GFP, green fluorescent protein; T+S, wild-type seedling containing target and silencer loci.
Figure 3
DNA methylation and RNA accumulation in the dms4 mutant. (A) DNA cytosine methylation (CG, black; CNG, blue; CNN, red; where N is A, T or C) is induced at the targeted enhancer (thick black bar) in wild-type plants (T+S) by primary siRNAs produced from the hairpin RNA trigger. Methylation triggered by Pol IV-dependent secondary siRNAs (Daxinger et al, 2009) is present at the downstream region (black line in white bar). In the dms4-1 mutant, methylation is reduced at the targeted enhancer and is nearly eliminated in the downstream region. (B) Histograms showing the abundance of siRNAs as measured in two sequencing by synthesis libraries, one each from wild-type (T+S) and dms4-1 mutant. The _y_-axis indicates the sum of the normalized abundance of each matching small RNA in TP2M. Top: a histogram of siRNA abundances matching the target enhancer region (primary siRNAs). Primary siRNAs (21, 22 and 24 nt) result from the redundant action of several DCL enzymes acting on the hairpin RNA trigger. RdDM is triggered by DCL3-generated 24 nt siRNAs (Daxinger et al, 2009). Bottom: a histogram of siRNA abundances matching in the 150 bp downstream from the target enhancer (secondary siRNAs). Pol IV-dependent 24 nt secondary siRNAs result from turnover of an enhancer-associated nascent RNA (Fig 1). The 24 nt primary siRNAs are reduced in abundance from 894 TP2M in wild type to 606 TP2M in the dms4-1 mutant, a decline of 32.2%; the 24 nt secondary siRNAs go from 702 TP2M in wild type to 31 TP2M in the dms4-1 mutant, a decline of 95.6%. (C) A Pol V-dependent transcript, IGN5 (Wierzbicki et al, 2008), is present in wild-type T+S plants, reduced in the dms4-1 mutant and eliminated in an nrpe1 mutant. By contrast, a Pol II-dependent solo LTR transcript (Wierzbicki et al, 2008) is reactivated in the two mutants owing to release of silencing. The nrpe1-12 allele was used in (C). DCL, DICER-LIKE; LTR, long terminal repeat; nrpe1, mutant defective in the largest subunits of Pol V; RT; reverse transcriptase; siRNA, small interfering RNA; T, wild-type plants containing the target locus; TP2M, transcripts per two million; T+S, wild-type plants containing target and silencer complexes.
Figure 4
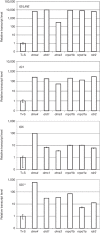
Transcription of RdDM targets in the dms4-1 mutant. Intergenic transcripts (IG/LINE, IG1, IG5 and IG5**) identified in the drd1 mutant (Huettel et al, 2006) were analysed by quantitative RT–PCR in dms4-1 plants and other mutants defective in RdDM (drd1, dms3, nrpe1, nrpd1 and rdr2; Kanno et al, 2008). A similar level of target de-repression was observed for all mutants. Transcription data are shown in a log scale. UPL7 (At3g53090) is the internal reference gene. Expression levels are relative to wild-type plants (T+S). Error bars, s.d.±mean. Error bars are not visible in some cases because of the log scale. RdDM, RNA-directed DNA methylation; RT–PCR, reverse transcriptase PCR; T+S, wild-type plants containing target and silencer constructs; UPL7, ubiquitin protein ligase 7.
Similar articles
- NRPD4, a protein related to the RPB4 subunit of RNA polymerase II, is a component of RNA polymerases IV and V and is required for RNA-directed DNA methylation.
He XJ, Hsu YF, Pontes O, Zhu J, Lu J, Bressan RA, Pikaard C, Wang CS, Zhu JK. He XJ, et al. Genes Dev. 2009 Feb 1;23(3):318-30. doi: 10.1101/gad.1765209. Genes Dev. 2009. PMID: 19204117 Free PMC article. - A conserved transcriptional regulator is required for RNA-directed DNA methylation and plant development.
He XJ, Hsu YF, Zhu S, Liu HL, Pontes O, Zhu J, Cui X, Wang CS, Zhu JK. He XJ, et al. Genes Dev. 2009 Dec 1;23(23):2717-22. doi: 10.1101/gad.1851809. Epub 2009 Nov 10. Genes Dev. 2009. PMID: 19903758 Free PMC article. - The RNA silencing enzyme RNA polymerase v is required for plant immunity.
López A, Ramírez V, García-Andrade J, Flors V, Vera P. López A, et al. PLoS Genet. 2011 Dec;7(12):e1002434. doi: 10.1371/journal.pgen.1002434. Epub 2011 Dec 29. PLoS Genet. 2011. PMID: 22242006 Free PMC article. - Molecular mechanisms of the RNA polymerases in plant RNA-directed DNA methylation.
Xie G, Du X, Hu H, Du J. Xie G, et al. Trends Biochem Sci. 2024 Mar;49(3):247-256. doi: 10.1016/j.tibs.2023.11.005. Epub 2023 Dec 9. Trends Biochem Sci. 2024. PMID: 38072749 Review. - RNA-Directed DNA Methylation: The Evolution of a Complex Epigenetic Pathway in Flowering Plants.
Matzke MA, Kanno T, Matzke AJ. Matzke MA, et al. Annu Rev Plant Biol. 2015;66:243-67. doi: 10.1146/annurev-arplant-043014-114633. Epub 2014 Dec 10. Annu Rev Plant Biol. 2015. PMID: 25494460 Review.
Cited by
- Interplay among RNA polymerases II, IV and V in RNA-directed DNA methylation at a low copy transgene locus in Arabidopsis thaliana.
You W, Lorkovic ZJ, Matzke AJ, Matzke M. You W, et al. Plant Mol Biol. 2013 May;82(1-2):85-96. doi: 10.1007/s11103-013-0041-4. Epub 2013 Mar 20. Plant Mol Biol. 2013. PMID: 23512103 Free PMC article. - Noncoding RNAs and DNA Methylation in Plants.
Zhao Y, Chen X. Zhao Y, et al. Natl Sci Rev. 2014 Jun;1(2):219-229. doi: 10.1093/nsr/nwu003. Natl Sci Rev. 2014. PMID: 25635229 Free PMC article. - Transgenerational maintenance of transgene body CG but not CHG and CHH methylation.
Dalakouras A, Dadami E, Zwiebel M, Krczal G, Wassenegger M. Dalakouras A, et al. Epigenetics. 2012 Sep;7(9):1071-8. doi: 10.4161/epi.21644. Epub 2012 Aug 6. Epigenetics. 2012. PMID: 22863736 Free PMC article. - Iwr1 protein is important for preinitiation complex formation by all three nuclear RNA polymerases in Saccharomyces cerevisiae.
Esberg A, Moqtaderi Z, Fan X, Lu J, Struhl K, Byström A. Esberg A, et al. PLoS One. 2011;6(6):e20829. doi: 10.1371/journal.pone.0020829. Epub 2011 Jun 10. PLoS One. 2011. PMID: 21695216 Free PMC article. - Reproductive developmental transcriptome analysis of Tripidium ravennae (Poaceae).
Maren N, Zhao F, Aryal R, Touchell D, Liu W, Ranney T, Ashrafi H. Maren N, et al. BMC Genomics. 2021 Jun 28;22(1):483. doi: 10.1186/s12864-021-07641-y. BMC Genomics. 2021. PMID: 34182921 Free PMC article.
References
- Aouida M, Pagé N, Leduc A, Peter M, Ramotar D (2004) A genome-wide screen in Saccharomyces cerevisiae reveals altered transport as a mechanism of resistance to the anticancer drug bleomycin. Cancer Res 64: 1102–1109 - PubMed
- Collins S et al. (2007) Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446: 806–810 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases