Structure of the MLL CXXC domain-DNA complex and its functional role in MLL-AF9 leukemia - PubMed (original) (raw)
Structure of the MLL CXXC domain-DNA complex and its functional role in MLL-AF9 leukemia
Tomasz Cierpicki et al. Nat Struct Mol Biol. 2010 Jan.
Abstract
The gene MLL (encoding the protein mixed-lineage leukemia) is the target of chromosomal translocations that cause leukemias with poor prognosis. All leukemogenic MLL fusion proteins retain the CXXC domain, which binds to nonmethylated CpG DNA sites. We present the solution structure of the MLL CXXC domain in complex with DNA, showing how the CXXC domain distinguishes nonmethylated from methylated CpG DNA. On the basis of the structure, we generated point mutations that disrupt DNA binding. Introduction of these mutations into the MLL-AF9 fusion protein resulted in increased DNA methylation of specific CpG nucleotides in Hoxa9, increased H3K9 methylation, decreased expression of Hoxa9-locus transcripts, loss of immortalization potential, and inability to induce leukemia in mice. These results establish that DNA binding by the CXXC domain and protection against DNA methylation is essential for MLL fusion leukemia. They also provide support for viewing this interaction as a potential target for therapeutic intervention.
Figures
Figure 1
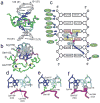
Structure of the CXXC domain – DNA complex and details of CpG recognition. a) representative conformer of the complex is shown in green (CpG binding loop 1182-1188 in magenta) and central 6 base pair region of DNA shown in blue and cyan; zinc atoms are shown as black spheres. b) Structure of the complex representing the view perpendicular to DNA axis; N- and C-terminal residues are labeled. Sidechains of Arg1150, Ser1152 and Leu1197 are shown in red. Cysteine resides coordinating zinc atoms are yellow. c) Schematic of protein-DNA contacts. Hydrogen bond, electrostatic, and van der Waals interactions of the protein backbone and sidechains with DNA are shown as red and blue arrows, respectively. The interaction of Arg1150 with minor groove is shown with green arrow. Ovals represent protein residues involved in base-specific (cyan) and electrostatic/van der Waals (green) contacts with DNA. d) Hydrogen bonds involving the carbonyls of Lys1185 and Lys1186 and N4-amine groups of Cyt106 and Cyt118. e) Hydrogen bonds formed between sidechains of Gln1187, Lys1186 and Gua107, Gua119, respectively. f) Close contacts between the protein backbone and cytosines in the CpG motif. The positions of H5 protons that are substituted by CH3 groups in methylated DNA are shown as gray spheres with the van der Waals radii of a methyl group. Backbone H and O atoms in intimate contact with the DNA are shown as magenta spheres with appropriate van der Waals radii.
Figure 2
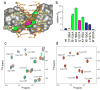
Design of mutations impairing interaction of the CXXC domain with DNA. a) Surface representation of the CXXC domain - DNA complex with indicated mutation sites. Residues involved in electrostatic interactions with DNA (Lys1185; Arg1154, Lys1193) are shown in green, Gln1187 forming hydrogen bond with guanine base in magenta, and Cys1188 located in close proximity to the DNA backbone in yellow. b) Relative dissociation constants (Kd) for binding of wildtype and mutant CXXC domains to DNApal determined using NMR titration. The colors of bars are the same as in panel a. Error bars indicate s.d. The Kd value for C1188D could not be determined due to very weak binding. The additional set of resides (shown in red) comprises mutation of N-terminal (Arg1150) and C-terminal (L1197 and M1200) residues to alanines; position of these residues on the structure of the complex (panel a) is omitted for clarity. c) Example of 15N-1H HSQC spectra showing titration of the Q1187A mutant (red) with increasing concentrations of DNApal (cyan - 1:1 protein-DNA ratio; blue - 1:4 ratio). d) Comparison of spectra of C1188D mutant without (red) and with DNApal in a 1:4 ratio (blue).
Figure 3
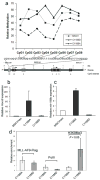
DNA binding activity of the MLL CXXC domain is required for MLL-AF9 to protect Hoxa9 from DNA methylation and induce Hoxa9 and mir-196b transcript expression, but not for binding to the locus. a) Relative methylation levels of CpGs in the upstream Hoxa9 locus in Mll null MEFs transfected with either MCSVneo, MSCVneo-MLL-AF9(C1188A), or MSCVneo-MLL-AF9(C1188D) and pSuper, after one week of puromycin selection. Bisulfite treatment, PCR and sequencing on genomic DNA samples were performed three times, and the results of one representative experiment are shown. Expression levels of Hoxa9 (b) and mir196b (c) in bone marrow progenitor cells transduced with MSCVneo vector, MLL-AF9(C1188A), or MLL-AF9(C1188D). Cells were harvested after one week culture in methylcellulose and expression levels of Hoxa9 and mir196b were quantified with real-time RT-PCR. Shown are average relative expression levels (s.d.). (d) ChIP assay performed on Phoenix cells transfected with FLAG tagged MSCV-MLL-AF9(C1188A) or MSCV-MLL-AF9(C1188D). Chromatin was immunoprecipitated with the indicated antibodies and real time PCR was performed with primers that localize near mir-196b in the upstream region of the HOXA9 locus. Samples were run in triplicate and were normalized to GAPDH and input chromatin, with error bars showing s.d.
Figure 4
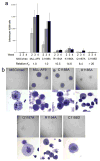
DNA binding activity of the MLL CXXC domain in MLL-AF9 is required for increased proliferative capacity and immortalization in a bone marrow progenitor serial replating assay. a) Average numbers of colonies for each of the four weeks after plating or re-plating in methylcellulose are shown for bone marrow progenitor cells expressing MLL-AF9 or MLL-AF9 with various CXXC domain point mutations, with error bars showing standard error. Also shown are the relative Kd values for binding of the wild type and mutated MLL CXXC domains to DNA. b) Digital photographs showing colony (above) and cell (below) morphologies of transduced bone marrow cells at the end of week 4 of the colony assay.
Figure 5
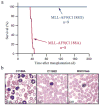
DNA binding activity of the MLL CXXC domain is required for MLL-AF9 to cause leukemia in vivo. a) Survival curve of mice transplanted with bone marrow progenitor cells infected with either MSCVneo-MLL-AF9(C1188A) or MSCVneo-MLL-AF9(C1188D). No mice transplanted with empty vector MSCVneo died (data not shown). b) Peripheral blood from mice at time of sacrifice (MLL-AF9(C1188A)) or at two months after bone marrow transplants (MSCVneo and MLL-AF9(C1188D)).
Figure 6
Model of the regulation of Hoxa9 locus transcription by the CXXC domain of MLL-AF9. a) The CXXC domain of MLL-AF9 protects specific CpG sequences within the Hoxa9 locus from methylation and maintains transcription within the locus; b) disruption of DNA binding function of CXXC domain by C1188D mutation results in increased methylation of the same CpGs, increased H3K9 trimethylation, and silencing of Hoxa9 and mir196b.
Similar articles
- Functional specificity of CpG DNA-binding CXXC domains in mixed lineage leukemia.
Risner LE, Kuntimaddi A, Lokken AA, Achille NJ, Birch NW, Schoenfelt K, Bushweller JH, Zeleznik-Le NJ. Risner LE, et al. J Biol Chem. 2013 Oct 11;288(41):29901-10. doi: 10.1074/jbc.M113.474858. Epub 2013 Aug 29. J Biol Chem. 2013. PMID: 23990460 Free PMC article. - Solution structure of the nonmethyl-CpG-binding CXXC domain of the leukaemia-associated MLL histone methyltransferase.
Allen MD, Grummitt CG, Hilcenko C, Min SY, Tonkin LM, Johnson CM, Freund SM, Bycroft M, Warren AJ. Allen MD, et al. EMBO J. 2006 Oct 4;25(19):4503-12. doi: 10.1038/sj.emboj.7601340. Epub 2006 Sep 21. EMBO J. 2006. PMID: 16990798 Free PMC article. - Binding to nonmethylated CpG DNA is essential for target recognition, transactivation, and myeloid transformation by an MLL oncoprotein.
Ayton PM, Chen EH, Cleary ML. Ayton PM, et al. Mol Cell Biol. 2004 Dec;24(23):10470-8. doi: 10.1128/MCB.24.23.10470-10478.2004. Mol Cell Biol. 2004. PMID: 15542854 Free PMC article. - Learning from mouse models of MLL fusion gene-driven acute leukemia.
Schwaller J. Schwaller J. Biochim Biophys Acta Gene Regul Mech. 2020 Aug;1863(8):194550. doi: 10.1016/j.bbagrm.2020.194550. Epub 2020 Apr 19. Biochim Biophys Acta Gene Regul Mech. 2020. PMID: 32320749 Review. - The molecular biology of mixed lineage leukemia.
Slany RK. Slany RK. Haematologica. 2009 Jul;94(7):984-93. doi: 10.3324/haematol.2008.002436. Epub 2009 Jun 16. Haematologica. 2009. PMID: 19535349 Free PMC article. Review.
Cited by
- Kdm2b maintains murine embryonic stem cell status by recruiting PRC1 complex to CpG islands of developmental genes.
He J, Shen L, Wan M, Taranova O, Wu H, Zhang Y. He J, et al. Nat Cell Biol. 2013 Apr;15(4):373-84. doi: 10.1038/ncb2702. Epub 2013 Mar 17. Nat Cell Biol. 2013. PMID: 23502314 Free PMC article. - Structural insights into inhibition of the bivalent menin-MLL interaction by small molecules in leukemia.
Shi A, Murai MJ, He S, Lund G, Hartley T, Purohit T, Reddy G, Chruszcz M, Grembecka J, Cierpicki T. Shi A, et al. Blood. 2012 Nov 29;120(23):4461-9. doi: 10.1182/blood-2012-05-429274. Epub 2012 Aug 30. Blood. 2012. PMID: 22936661 Free PMC article. - Insights in dynamic kinome reprogramming as a consequence of MEK inhibition in MLL-rearranged AML.
Kampen KR, Ter Elst A, Mahmud H, Scherpen FJ, Diks SH, Peppelenbosch MP, de Haas V, Guryev V, de Bont ES. Kampen KR, et al. Leukemia. 2014 Mar;28(3):589-99. doi: 10.1038/leu.2013.342. Epub 2013 Nov 18. Leukemia. 2014. PMID: 24240200 - Crystal structure of the N-terminal region of human Ash2L shows a winged-helix motif involved in DNA binding.
Chen Y, Wan B, Wang KC, Cao F, Yang Y, Protacio A, Dou Y, Chang HY, Lei M. Chen Y, et al. EMBO Rep. 2011 Jun 10;12(8):797-803. doi: 10.1038/embor.2011.101. EMBO Rep. 2011. PMID: 21660059 Free PMC article. - RNAi-mediated silencing of MLL-AF9 reveals leukemia-associated downstream targets and processes.
Fleischmann KK, Pagel P, Schmid I, Roscher AA. Fleischmann KK, et al. Mol Cancer. 2014 Feb 11;13:27. doi: 10.1186/1476-4598-13-27. Mol Cancer. 2014. PMID: 24517546 Free PMC article.
References
- Tkachuk DC, Kohler S, Cleary ML. Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell. 1992;71:691–700. - PubMed
- Sedkov Y, Tillib S, Mizrokhi L, Mazo A. The bithorax complex is regulated by trithorax earlier during Drosophila embryogenesis than is the Antennapedia complex, correlating with a bithorax-like expression pattern of distinct early trithorax transcripts. Development. 1994;120:1907–17. - PubMed
- Yu BD, Hess JL, Horning SE, Brown GA, Korsmeyer SJ. Altered Hox expression and segmental identity in Mll-mutant mice. Nature. 1995;378:505–8. - PubMed
- Muller J, Gaunt S, Lawrence PA. Function of the Polycomb protein is conserved in mice and flies. Development. 1995;121:2847–52. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 AI007508/AI/NIAID NIH HHS/United States
- CA105049/CA/NCI NIH HHS/United States
- R01 HL087188/HL/NHLBI NIH HHS/United States
- P01 CA105049/CA/NCI NIH HHS/United States
- HL087188/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases