Exome sequencing of a multigenerational human pedigree - PubMed (original) (raw)
. 2009 Dec 14;4(12):e8232.
doi: 10.1371/journal.pone.0008232.
Dan Burges, Eric Powell, Cherylyn Almonte, Jia Huang, Stuart Young, Benjamin Boese, Mike Schmidt, Margaret A Pericak-Vance, Eden Martin, Xinmin Zhang, Timothy T Harkins, Stephan Züchner
Affiliations
- PMID: 20011588
- PMCID: PMC2788131
- DOI: 10.1371/journal.pone.0008232
Exome sequencing of a multigenerational human pedigree
Dale J Hedges et al. PLoS One. 2009.
Erratum in
- PLoS One. 2009;4(12). doi: 10.1371/annotation/b0fe9dd5-16e1-4b50-b590-263518fbd5eb. Hedges, Dale [corrected to Hedges, Dale J]
Abstract
Over the next few years, the efficient use of next-generation sequencing (NGS) in human genetics research will depend heavily upon the effective mechanisms for the selective enrichment of genomic regions of interest. Recently, comprehensive exome capture arrays have become available for targeting approximately 33 Mb or approximately 180,000 coding exons across the human genome. Selective genomic enrichment of the human exome offers an attractive option for new experimental designs aiming to quickly identify potential disease-associated genetic variants, especially in family-based studies. We have evaluated a 2.1 M feature human exome capture array on eight individuals from a three-generation family pedigree. We were able to cover up to 98% of the targeted bases at a long-read sequence read depth of > or = 3, 86% at a read depth of > or = 10, and over 50% of all targets were covered with > or = 20 reads. We identified up to 14,284 SNPs and small indels per individual exome, with up to 1,679 of these representing putative novel polymorphisms. Applying the conservative genotype calling approach HCDiff, the average rate of detection of a variant allele based on Illumina 1 M BeadChips genotypes was 95.2% at > or = 10x sequence. Further, we propose an advantageous genotype calling strategy for low covered targets that empirically determines cut-off thresholds at a given coverage depth based on existing genotype data. Application of this method was able to detect >99% of SNPs covered > or = 8x. Our results offer guidance for "real-world" applications in human genetics and provide further evidence that microarray-based exome capture is an efficient and reliable method to enrich for chromosomal regions of interest in next-generation sequencing experiments.
Conflict of interest statement
Competing Interests: Roche provided complementary reagents and services for conducting some components of this study. The academic members of this study have no conflicting financial interests to declare, such as consulting fees, employment with a commercial partner, or similar. The manuscript was prepared solely under the direction of the academic authors. They adhere to all PLoS ONE policies on sharing data and materials, as detailed online in the guide for authors http://www.plosone.org/static/policies.action#sharing. Authors Benjamin Boese, Xinmin Zhang, and Timothy Harkins are employed by Roche, Inc., which develops and sells next generation sequencing and genome capture technology.
Figures
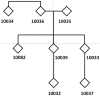
Figure 1. Studied three-generational pedigree.
Pedigree of eight individuals of European descent that was studied with exome capture arrays.
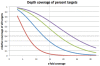
Figure 2. Sequence coverage of targeted exons.
The graph illustrates the cumulative coverage of targeted bases after sequencing 0.5 Gbp (red), 1 Gbp (blue), 1.5 Gbp (green), and 2 Gbp (purple). 1 Gb resulted in nearly 10x coverage of 50% of all targets; 2 Gb of data increase this number to 88%. Depending on a studies goal, maximum coverage might not always be required.
Figure 3. Estimated error rates.
Sensitivity of genotype calling based on HCDiff SNPs, AllDiff SNPs, and the proposed coverage-dependent genotype calling approach. A) False negative rates are based on concordance with a subset of 44,513 SNPs that overlapped with genotypes obtained with Illumina 1 M Duo BeadChips. The coverage-dependent variant calling approach that calibrates cut-off rates according to array-based genotypes is the most sensitive method, detecting >96% of SNPs at 5x coverage and >99% of all SNPs at ≥8x coverage. B) False positive rates. HCDiff is the most conservative algorithm, resulting in a smaller false positive rate, while the more relaxed dynamic genotype calling algorithm results in twice as high error rates at lower coverage.
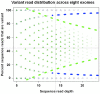
Figure 4. Variant read distribution across eight exomes.
Illustration of the dynamic nature of optimal cut-off rates for calling heterozygous/homozygous variants. At lower coverage (<10x) the ideal cut-off is 88% variant reads in our data, while it is 78% at coverage ≥20. Optimal usage of data should take advantage even of low covered targets. Data are based on comparison to Illumina genotyped SNPs. Green triangles: Illumina heterozygous genotypes, Blue diamonds: Illumina homozygous genotypes. NGS genotypes are placed according to their percent variant reads (y axis).
Similar articles
- Exome sequencing reveals comprehensive genomic alterations across eight cancer cell lines.
Chang H, Jackson DG, Kayne PS, Ross-Macdonald PB, Ryseck RP, Siemers NO. Chang H, et al. PLoS One. 2011;6(6):e21097. doi: 10.1371/journal.pone.0021097. Epub 2011 Jun 20. PLoS One. 2011. PMID: 21701589 Free PMC article. - Comparison of solution-based exome capture methods for next generation sequencing.
Sulonen AM, Ellonen P, Almusa H, Lepistö M, Eldfors S, Hannula S, Miettinen T, Tyynismaa H, Salo P, Heckman C, Joensuu H, Raivio T, Suomalainen A, Saarela J. Sulonen AM, et al. Genome Biol. 2011 Sep 28;12(9):R94. doi: 10.1186/gb-2011-12-9-r94. Genome Biol. 2011. PMID: 21955854 Free PMC article. - Evaluation of whole-genome sequencing of four Chinese crested dogs for variant detection using the ion proton system.
Viluma A, Sayyab S, Mikko S, Andersson G, Bergström TF. Viluma A, et al. Canine Genet Epidemiol. 2015 Oct 8;2:16. doi: 10.1186/s40575-015-0029-2. eCollection 2015. Canine Genet Epidemiol. 2015. PMID: 26457193 Free PMC article. - Novel genomic techniques open new avenues in the analysis of monogenic disorders.
Kuhlenbäumer G, Hullmann J, Appenzeller S. Kuhlenbäumer G, et al. Hum Mutat. 2011 Feb;32(2):144-51. doi: 10.1002/humu.21400. Hum Mutat. 2011. PMID: 21280146 Review. - Finding the lost treasures in exome sequencing data.
Samuels DC, Han L, Li J, Quanghu S, Clark TA, Shyr Y, Guo Y. Samuels DC, et al. Trends Genet. 2013 Oct;29(10):593-9. doi: 10.1016/j.tig.2013.07.006. Epub 2013 Aug 22. Trends Genet. 2013. PMID: 23972387 Free PMC article. Review.
Cited by
- New approaches for the discovery of lipid‑related genes.
Curran JE, Meikle PJ, Blangero J. Curran JE, et al. Clin Lipidol. 2011;6(5):495-500. Epub 2017 Jan 18. Clin Lipidol. 2011. PMID: 32983264 Free PMC article. - Exploration of Mutated Genes and Prediction of Potential Biomarkers for Childhood-Onset Schizophrenia Using an Integrated Bioinformatic Analysis.
He F, Zhou YM, Qi YJ, Huang HH, Guan L, Luo J, Cheng YH, Zheng Y. He F, et al. Front Aging Neurosci. 2022 Jun 16;14:829217. doi: 10.3389/fnagi.2022.829217. eCollection 2022. Front Aging Neurosci. 2022. PMID: 35783128 Free PMC article. - Another locus, a new method.
Singleton AB, Gibbs JR. Singleton AB, et al. Brain. 2010 Dec;133(Pt 12):3492-3. doi: 10.1093/brain/awq331. Brain. 2010. PMID: 21126992 Free PMC article. No abstract available. - Exome sequencing reveals comprehensive genomic alterations across eight cancer cell lines.
Chang H, Jackson DG, Kayne PS, Ross-Macdonald PB, Ryseck RP, Siemers NO. Chang H, et al. PLoS One. 2011;6(6):e21097. doi: 10.1371/journal.pone.0021097. Epub 2011 Jun 20. PLoS One. 2011. PMID: 21701589 Free PMC article. - Positional cloning and next-generation sequencing identified a TGM6 mutation in a large Chinese pedigree with acute myeloid leukaemia.
Pan LL, Huang YM, Wang M, Zhuang XE, Luo DF, Guo SC, Zhang ZS, Huang Q, Lin SL, Wang SY. Pan LL, et al. Eur J Hum Genet. 2015 Feb;23(2):218-23. doi: 10.1038/ejhg.2014.67. Epub 2014 Apr 23. Eur J Hum Genet. 2015. PMID: 24755948 Free PMC article.
References
- Taly V, Kelly BT, Griffiths AD. Droplets as microreactors for high-throughput biology. Chembiochem. 2007;8(3):263–272. - PubMed
- Hodges E, Xuan Z, Balija V, Kramer M, Molla MN, et al. Genome-wide in situ exon capture for selective resequencing. Nat Genet. 2007;39(12):1522–1527. - PubMed
Publication types
MeSH terms
Grants and funding
- P01 NS026630/NS/NINDS NIH HHS/United States
- U54 NS065712/NS/NINDS NIH HHS/United States
- RC2 HG005605/HG/NHGRI NIH HHS/United States
- NS065712/NS/NINDS NIH HHS/United States
- RC2HG005605/HG/NHGRI NIH HHS/United States
- NS026630/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous