Mutations in the formin gene INF2 cause focal segmental glomerulosclerosis - PubMed (original) (raw)
Mutations in the formin gene INF2 cause focal segmental glomerulosclerosis
Elizabeth J Brown et al. Nat Genet. 2010 Jan.
Erratum in
- Nat Genet. 2010 Apr;42(4):361. Tonna, Stephen J [added]
Abstract
Focal segmental glomerulosclerosis (FSGS) is a pattern of kidney injury observed either as an idiopathic finding or as a consequence of underlying systemic conditions. Several genes have been identified that, when mutated, lead to inherited FSGS and/or the related nephrotic syndrome. These findings have accelerated the understanding of glomerular podocyte function and disease, motivating our search for additional FSGS genes. Using linkage analysis, we identified a locus for autosomal-dominant FSGS susceptibility on a region of chromosome 14q. By sequencing multiple genes in this region, we detected nine independent nonconservative missense mutations in INF2, which encodes a member of the formin family of actin-regulating proteins. These mutations, all within the diaphanous inhibitory domain of INF2, segregate with FSGS in 11 unrelated families and alter highly conserved amino acid residues. The observation that alterations in this podocyte-expressed formin cause FSGS emphasizes the importance of fine regulation of actin polymerization in podocyte function.
Figures
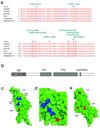
Figure 1. Pedigrees
Pedigree diagrams of families FGBR and FGJN. Pedigree identifier and the specific mutations segregating in each family are shown.
Figure 2. INF2 mutations
a. Disease-segregating INF2 mutations shown aligned with wild-type INF2 protein sequence from humans, chimpanzee, mouse, rat, opossum, and zebrafish. All of these disease mutations occur in evolutionarily conserved residues within the DID. b. Schematic showing INF2 protein domain structure and location of mutations. c, d, and e: Model of mouse INF2 amino acids 1–330, based on the structure of mDia1 (1). Mutated residues are shown in red, and residues important for the interaction with DAD are shown in blue. c. View of mDia1 showing the positions of A13 and R218 (red). Residues important for the direct interaction with DAD are shown in blue, including R106 (corresponding to K213 in mDia1), N110 (corresponding to N217 in mDia1), A149 (corresponding to A256 in mDia1), and I152 (corresponding to I259 in mDia1). Based on the crystal structure of the mDia1 DID/DAD complex (reference 22), the alpha helical INF2 DAD is predicted to lie in the pocket containing these residues, with its N-terminus (D974) contacting R106 and N110, and L986 contacting A149 and I152. In this model, we predict that R218 would contact residues C-terminal to L986. d. Close-up of the portion of the INF2 region predicted to interact with the DAD. e. 180 degree rotation of the structure shown in F1, showing L42, S186, and E220.
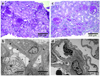
Figure 3. Histopathology
a, b: Kidney biopsy findings in an affected member of family FSB with an E220K INF2 mutation. This patient was 21 years of age at the time of biopsy with an estimated glomerular filtration rate (GFR) of 96 ml/min/1.73m2. She had 3+ urine protein and no hematuria. She developed ESRD seven years after the biopsy was performed. a. Light micrograph (PAS) showing focal global and segmental glomerulosclerosis. b. Electron micrograph showing segmental foot process effacement in some loops (arrowheads) and focally irregular morphology of preserved foot processes. Inset: Higher magnification electron micrograph showing foot processes en face, projecting from a major process. The foot processes appear irregular and jagged, often with prominent longitudinal actin bundles. c, d: Kidney biopsy findings from a different INF2 mutant patient with an R218Q mutation, from family FGJN. This patient was 26 years old at the time of biopsy. Estimated GFR was 46 ml/min/1.73m2 and urine showed 3+ protein and trace blood at the time. c: Light micrograph (PAS) showing focal and segmental glomerulosclerosis, with moderate chronic parenchymal damage. d: Electron micrograph showing segmental foot process effacement (arrowheads).
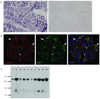
Figure 4. INF2 expression
a. RNA in situ hybridization in adult human kidney with digoxigenin-labeled probe targeted against INF2 mRNA, followed by immunohistochemical staining with labeled antibody to digoxigenin. INF2 mRNA expression is apparent in podocytes as well as some tubule cells. Sense control experiment is shown on the right. b. Immunofluoresence staining of mouse kidney with DAPI (blue) and antibodies against nephrin (red) and INF2 (green). INF2 staining is observed in the glomeruli in an epithelial cell pattern. Arrows show areas of nephrin and INF2 colocalization. Arrowhead shows a cell expressing INF2 but not nephrin. d. Northern blot. Gel lanes: 1, Brain; 2, Placenta; 3, Skeletal muscle; 4, Heart; 5, Kidney; 6, Pancreas; 7, Liver; 8, Lung; 9, Spleen; 10, Colon.
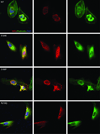
Figure 5. Expression of INF2 variants
Localization of INF2 variants in undifferentiated cultured human podocytes. Cells were cultured on coverslips coated with type I collagen. INF2 is visualized using a Cy3 conjugated antibody against its C-terminus . FITC phalloidin was used to stain F-actin, and nuclei are stained with DAPI. First column: cells expressing wild-type and three different mutant forms of INF2. Second column: INF2 channel. Third column: Phalloidin. Rows: results for WT and three mutant forms of INF2: E184K, S186P, and R218Q. E184K and R218Q mutants show diffuse localization of INF2 and F-actin compared with the perinuclear INF2 staining seen with the WT expressing cells. The S186P looked similar to WT expressing cells, but INF2 expression was more vermiform in appearance. In all three mutants, stress fibers and cortical actin appeared to be less prominent.
Similar articles
- Inverted formin 2 mutations with variable expression in patients with sporadic and hereditary focal and segmental glomerulosclerosis.
Gbadegesin RA, Lavin PJ, Hall G, Bartkowiak B, Homstad A, Jiang R, Wu G, Byrd A, Lynn K, Wolfish N, Ottati C, Stevens P, Howell D, Conlon P, Winn MP. Gbadegesin RA, et al. Kidney Int. 2012 Jan;81(1):94-9. doi: 10.1038/ki.2011.297. Epub 2011 Aug 24. Kidney Int. 2012. PMID: 21866090 Free PMC article. - Mutations in INF2 are a major cause of autosomal dominant focal segmental glomerulosclerosis.
Boyer O, Benoit G, Gribouval O, Nevo F, Tête MJ, Dantal J, Gilbert-Dussardier B, Touchard G, Karras A, Presne C, Grunfeld JP, Legendre C, Joly D, Rieu P, Mohsin N, Hannedouche T, Moal V, Gubler MC, Broutin I, Mollet G, Antignac C. Boyer O, et al. J Am Soc Nephrol. 2011 Feb;22(2):239-45. doi: 10.1681/ASN.2010050518. Epub 2011 Jan 21. J Am Soc Nephrol. 2011. PMID: 21258034 Free PMC article. - Variable renal phenotype in a family with an INF2 mutation.
Lee HK, Han KH, Jung YH, Kang HG, Moon KC, Ha IS, Choi Y, Cheong HI. Lee HK, et al. Pediatr Nephrol. 2011 Jan;26(1):73-6. doi: 10.1007/s00467-010-1644-5. Epub 2010 Aug 28. Pediatr Nephrol. 2011. PMID: 20803156 - Role of formin INF2 in human diseases.
Zhao Y, Zhang H, Wang H, Ye M, Jin X. Zhao Y, et al. Mol Biol Rep. 2022 Jan;49(1):735-746. doi: 10.1007/s11033-021-06869-x. Epub 2021 Oct 26. Mol Biol Rep. 2022. PMID: 34698992 Review. - The formin INF2 in disease: progress from 10 years of research.
Labat-de-Hoz L, Alonso MA. Labat-de-Hoz L, et al. Cell Mol Life Sci. 2020 Nov;77(22):4581-4600. doi: 10.1007/s00018-020-03550-7. Epub 2020 May 25. Cell Mol Life Sci. 2020. PMID: 32451589 Free PMC article. Review.
Cited by
- INF2 mutations cause kidney disease through a gain-of-function mechanism.
Subramanian B, Williams S, Karp S, Hennino MF, Jacas S, Lee M, Riella CV, Alper SL, Higgs HN, Pollak MR. Subramanian B, et al. Sci Adv. 2024 Nov 15;10(46):eadr1017. doi: 10.1126/sciadv.adr1017. Epub 2024 Nov 13. Sci Adv. 2024. PMID: 39536114 Free PMC article. - Mechanisms of podocyte injury in genetic kidney disease.
Mann N, Sun H, Majmundar AJ. Mann N, et al. Pediatr Nephrol. 2024 Nov 1. doi: 10.1007/s00467-024-06551-x. Online ahead of print. Pediatr Nephrol. 2024. PMID: 39485497 Review. - Altered Endoplasmic Reticulum Integrity and Organelle Interactions in Living Cells Expressing INF2 Variants.
Tran QTH, Kondo N, Ueda H, Matsuo Y, Tsukaguchi H. Tran QTH, et al. Int J Mol Sci. 2024 Sep 10;25(18):9783. doi: 10.3390/ijms25189783. Int J Mol Sci. 2024. PMID: 39337270 Free PMC article. - Frameshift Mutation in PAX2 Related to Focal Segmental Glomerular Sclerosis: A Case Report and Literature Review.
Hu X, Lin W, Luo Z, Zhong Y, Xiao X, Tang R. Hu X, et al. Mol Genet Genomic Med. 2024 Sep;12(9):e70006. doi: 10.1002/mgg3.70006. Mol Genet Genomic Med. 2024. PMID: 39235128 Free PMC article. Review. - Actin Cytoskeleton and Integrin Components Are Interdependent for Slit Diaphragm Maintenance in Drosophila Nephrocytes.
Delaney M, Zhao Y, van de Leemput J, Lee H, Han Z. Delaney M, et al. Cells. 2024 Aug 14;13(16):1350. doi: 10.3390/cells13161350. Cells. 2024. PMID: 39195240 Free PMC article.
References
- Kaplan JM, et al. Mutations in ACTN4, encoding alpha-actinin-4, cause familial focal segmental glomerulosclerosis. Nat Genet. 2000;24:251–256. - PubMed
- Winn MP, et al. A mutation in the TRPC6 cation channel causes familial focal segmental glomerulosclerosis. Science. 2005;308:1801–1804. - PubMed
- Chhabra ES, Higgs HN. INF2 Is a WASP homology 2 motif-containing formin that severs actin filaments and accelerates both polymerization and depolymerization. J Biol Chem. 2006;281:26754–26767. - PubMed
- D'Agati VD, Fogo AB, Bruijn JA, Jennette JC. Pathologic classification of focal segmental glomerulosclerosis: a working proposal. Am J Kidney Dis. 2004;43:368–382. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 DK054931/DK/NIDDK NIH HHS/United States
- DK54931/DK/NIDDK NIH HHS/United States
- R01 DK054931-10A1/DK/NIDDK NIH HHS/United States
- R56 DK054931/DK/NIDDK NIH HHS/United States
- DK073091/DK/NIDDK NIH HHS/United States
- K08 DK073091/DK/NIDDK NIH HHS/United States
- R01 GM069818/GM/NIGMS NIH HHS/United States
- GM069818/GM/NIGMS NIH HHS/United States
- K08 DK080947/DK/NIDDK NIH HHS/United States
- R01 DK054931-13/DK/NIDDK NIH HHS/United States
- DK080947/DK/NIDDK NIH HHS/United States
- R01 DK088826/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases