Enhancers: the abundance and function of regulatory sequences beyond promoters - PubMed (original) (raw)
Review
Enhancers: the abundance and function of regulatory sequences beyond promoters
Michael Bulger et al. Dev Biol. 2010.
Abstract
Transcriptional control in mammals and Drosophila is often mediated by regulatory sequences located far from gene promoters. Different classes of such elements - particularly enhancers, but also locus control regions and insulators - have been defined by specific functional assays, although it is not always clear how these assays relate to the function of these elements within their native loci. Recent advances in genomics suggest, however, that such elements are highly abundant within the genome and may represent the primary mechanism by which cell- and developmental-specific gene expression is accomplished. In this review, we discuss the functional parameters of enhancers as defined by specific assays, along with the frequency with which they occur in the genome. In addition, we examine the available evidence for the mechanism by which such elements communicate or interact with the promoters they regulate.
Copyright 2010 Elsevier Inc. All rights reserved.
Figures
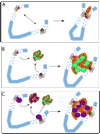
Figure 1
Potential mechanisms underlying distal enhancer-promoter colocalization. A. The “traditional” view of enhancer-promoter interactions, in which proteins bound to the enhancer (ENH) and other proteins bound to the promoter (PRO) directly interact with each other to facilitate transcriptional activation, whether by recruitment of RNA polymerase II (as shown), stimulation of RNA polymerase II elongation (as demonstrated for the β-globin LCR), or another mechanism. Blue circles represent nucleosomes; based on the best available evidence, we have depicted the majority of intervening sequence between the enhancer and promoter as condensed to the 30 nm fiber, although higher-order chromatin structure is not well understood and so compaction may be more extensive than this. B. Enhancer-promoter colocalization by association with RNA polymerase II transcription “factories”. In this model, factors bound to both enhancer and promoter independently recruit RNA polymerase II; in a nuclear environment consisting of transcription “factories”, however, this amounts to enhancer and promoter colocalizing to the same “factory”. The green circles represent as-yet undefined, non-Pol II proteins presumed to be present in transcription factories. C. Enhancer and promoter interactions with specific transcription factories. In this model, different genes associate with different kinds of transcription factories. Such specific association is presumably mediated by some common factor or complex recruited to both the enhancer and the promoter (denoted by violet or green circles, respectively), and would be expected to underlie specific interchromosomal interactions.
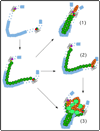
Figure 2
Potential role of intervening DNA between enhancers and promoters, and factors that associate with it, in enhancer function. An enhancer and a promoter within a gene locus (top left) are both bound by sequence-specific factors, which in turn serve to recruit a factor or complex that then associates with chromatin throughout the locus (bottom left). We depict several potential mechanisms that might then ensue: (1) factors bound between the enhancer and promoter serve to organize the intervening chromatin in order to bring the two elements together spatially; (2) factors extending from the enhancer to the promoter along the intervening chromatin serve as the primary signal for gene activation, with no necessary role for enhancer-promoter juxtaposition; (3) factors associating with the intervening chromatin organize the locus to bring both enhancer and promoter to a transcription factory and accomplish gene activation. Conceivably, enhancer-promoter juxtaposition at transcription factories may be an event secondary to gene activation, as in (2).
Similar articles
- Modulation of enhancer-promoter interactions by insulators in the Drosophila embryo.
Cai H, Levine M. Cai H, et al. Nature. 1995 Aug 10;376(6540):533-6. doi: 10.1038/376533a0. Nature. 1995. PMID: 7637789 - Effects of cis arrangement of chromatin insulators on enhancer-blocking activity.
Cai HN, Shen P. Cai HN, et al. Science. 2001 Jan 19;291(5503):493-5. doi: 10.1126/science.291.5503.493. Science. 2001. PMID: 11161205 - Loss of insulator activity by paired Su(Hw) chromatin insulators.
Muravyova E, Golovnin A, Gracheva E, Parshikov A, Belenkaya T, Pirrotta V, Georgiev P. Muravyova E, et al. Science. 2001 Jan 19;291(5503):495-8. doi: 10.1126/science.291.5503.495. Science. 2001. PMID: 11161206 - Evaluating Enhancer Function and Transcription.
Field A, Adelman K. Field A, et al. Annu Rev Biochem. 2020 Jun 20;89:213-234. doi: 10.1146/annurev-biochem-011420-095916. Epub 2020 Mar 20. Annu Rev Biochem. 2020. PMID: 32197056 Review. - Regulatory Landscaping: How Enhancer-Promoter Communication Is Sculpted in 3D.
Robson MI, Ringel AR, Mundlos S. Robson MI, et al. Mol Cell. 2019 Jun 20;74(6):1110-1122. doi: 10.1016/j.molcel.2019.05.032. Mol Cell. 2019. PMID: 31226276 Review.
Cited by
- TENET 2.0: Identification of key transcriptional regulators and enhancers in lung adenocarcinoma.
Mullen DJ, Yan C, Kang DS, Zhou B, Borok Z, Marconett CN, Farnham PJ, Offringa IA, Rhie SK. Mullen DJ, et al. PLoS Genet. 2020 Sep 14;16(9):e1009023. doi: 10.1371/journal.pgen.1009023. eCollection 2020 Sep. PLoS Genet. 2020. PMID: 32925947 Free PMC article. - Intestinal master transcription factor CDX2 controls chromatin access for partner transcription factor binding.
Verzi MP, Shin H, San Roman AK, Liu XS, Shivdasani RA. Verzi MP, et al. Mol Cell Biol. 2013 Jan;33(2):281-92. doi: 10.1128/MCB.01185-12. Epub 2012 Nov 5. Mol Cell Biol. 2013. PMID: 23129810 Free PMC article. - Spirits in the Material World: Enhancer RNAs in Transcriptional Regulation.
Hou TY, Kraus WL. Hou TY, et al. Trends Biochem Sci. 2021 Feb;46(2):138-153. doi: 10.1016/j.tibs.2020.08.007. Epub 2020 Sep 1. Trends Biochem Sci. 2021. PMID: 32888773 Free PMC article. Review. - A comprehensive enhancer screen identifies TRAM2 as a key and novel mediator of YAP oncogenesis.
Li L, Ugalde AP, Scheele CLGJ, Dieter SM, Nagel R, Ma J, Pataskar A, Korkmaz G, Elkon R, Chien MP, You L, Su PR, Bleijerveld OB, Altelaar M, Momchev L, Manber Z, Han R, van Breugel PC, Lopes R, Ten Dijke P, van Rheenen J, Agami R. Li L, et al. Genome Biol. 2021 Jan 29;22(1):54. doi: 10.1186/s13059-021-02272-8. Genome Biol. 2021. PMID: 33514403 Free PMC article. - Transcriptionally active enhancers in human cancer cells.
Lidschreiber K, Jung LA, von der Emde H, Dave K, Taipale J, Cramer P, Lidschreiber M. Lidschreiber K, et al. Mol Syst Biol. 2021 Jan;17(1):e9873. doi: 10.15252/msb.20209873. Mol Syst Biol. 2021. PMID: 33502116 Free PMC article.
References
- Banerji J, Rusconi S, Schaffner W. Expression of a beta-globin gene is enhancer by remote SV40 DNA sequences. Cell. 1981;27:299–308. - PubMed
- Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. - PubMed
- Bender MA, Bulger M, Close J, Groudine M. Beta-globin gene switching and DNaseI sensitivity of the endogenous beta-globin locus in mice do not require the locus control region. Mol. Cell. 2000;5:387–393. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 DK070687/DK/NIDDK NIH HHS/United States
- R01 DK070687-04/DK/NIDDK NIH HHS/United States
- R01 HL065440/HL/NHLBI NIH HHS/United States
- R37 DK044746/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases