Conserved long noncoding RNAs transcriptionally regulated by Oct4 and Nanog modulate pluripotency in mouse embryonic stem cells - PubMed (original) (raw)
Conserved long noncoding RNAs transcriptionally regulated by Oct4 and Nanog modulate pluripotency in mouse embryonic stem cells
Jameelah Sheik Mohamed et al. RNA. 2010 Feb.
Abstract
The genetic networks controlling stem cell identity are the focus of intense interest, due to their obvious therapeutic potential as well as exceptional relevance to models of early development. Genome-wide mapping of transcriptional networks in mouse embryonic stem cells (mESCs) reveals that many endogenous noncoding RNA molecules, including long noncoding RNAs (lncRNAs), may play a role in controlling the pluripotent state. We performed a genome-wide screen that combined full-length mESC transcriptome genomic mapping data with chromatin immunoprecipitation genomic location maps of the key mESC transcription factors Oct4 and Nanog. We henceforth identified four mESC-expressed, conserved lncRNA-encoding genes residing proximally to active genomic binding sites of Oct4 and Nanog. Accordingly, these four genes have potential roles in pluripotency. We show that two of these lncRNAs, AK028326 (Oct4-activated) and AK141205 (Nanog-repressed), are direct targets of Oct4 and Nanog. Most importantly, we demonstrate that these lncRNAs are not merely controlled by mESC transcription factors, but that they themselves regulate developmental state: knockdown and overexpression of these transcripts lead to robust changes in Oct4 and Nanog mRNA levels, in addition to alterations in cellular lineage-specific gene expression and in the pluripotency of mESCs. We further characterize AK028326 as a co-activator of Oct4 in a regulatory feedback loop. These results for the first time implicate lncRNAs in the modulation of mESC pluripotency and expand the established mESC regulatory network model to include functional lncRNAs directly controlled by key mESC transcription factors.
Figures
FIGURE 1.
Graphical representation of the lncRNA genomic loci and approximate sites of transcription factor interaction. (Black boxes) Exonic sequences, commencing at the transcriptional start site (right-angle arrow); (gray ovals) putative Oct4 binding sites supported by Oct4 ChIP-PET data; (gray triangles) putative Nanog binding sites supported by Nanog ChIP-PET data; (small arrowheads) forward and reverse primer binding sites for real-time PCR primers. Gene length is indicated by the scale bar provided.
FIGURE 2.
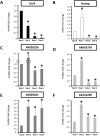
Retinoic acid (RA) treatment simultaneously induces mESC differentiation and impacts lncRNA transcription. (A) Oct4 expression was highest in undifferentiated mESCs and rapidly decreased over the 6-d time course. (B) Nanog expression was initially elevated on RA treatment but was rapidly down-regulated by days 4 and 6. The expression of putative Oct4-regulated lncRNAs (C,E: dark gray bars) and putative Nanog-regulated lncRNAs (D,F: light gray bars) was examined in differentiating mESCs. Transcription of the lncRNAs AK028326 (C) and AK043754 (D) was elevated by day 2 and down-regulated relative to undifferentiated mESCs by days 4 and 6. In contrast, transcription of AK005651 (E) and, to a lesser extent, AK141205 (F) remained elevated relative to undifferentiated mESCs at all time points examined. (*) Significant difference relative to undifferentiated mESCs (P < 0.05, N = 6 replicates).
FIGURE 3.
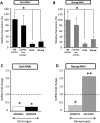
Differential expression of lncRNA upon robust Oct4 and Nanog RNAi knockdown. (A) shRNA-directed knockdown resulted in greater than or ∼80% reduction of Oct4 mRNA and Nanog mRNA, respectively, compared with the non-silencing control (NS control) and the pSUPER-PURO vector-only control (pSUPER-PURO control), 3 d post-transfection. (B) shRNA-directed knockdown resulted in comparable levels of Nanog mRNA reduction and Oct4 mRNA compared with the NS control and the pSUPER-NEO vector-only control (pSUPER-NEO control), 3 d post-transfection. (C) Oct4 RNAi resulted in down-regulation of the two putative Oct4 targets, AK005651 and AK028326. (D) Nanog RNAi resulted in down-regulation of the putative Nanog target AK043754 and up-regulation of AK141205. (Asterisks) Significant difference from control samples (*, P < 0.05; **, P < 0.01) relative to the NS control (dotted black line).
FIGURE 4.
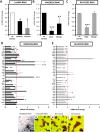
Expression of lncRNAs, pluripotent markers, trophectoderm markers, and epiblast lineage markers in response to lncRNA silencing (RNAi). (A) AK028326 and AK141205 are significantly down-regulated in response to siRNA treatment. (B) AK028326 RNAi was associated with significant down-regulation of both Oct4 and Nanog. (C) AK141205 RNAi was associated with significant down-regulation of Oct4, but Nanog levels remained unchanged. (D) In AK028326 siRNA-treated mESCs, there was a significant enhancement of the mesodermal marker T/Brachyury and the trophectoderm markers Cdx2, Hand1, Gata3, and Eomes; down-regulation of pluripotent markers; and unchanged levels of the housekeeping genes Gapdh and Ahcy. (E) In AK141205 siRNA-treated mESCs, there was general down-regulation of the pluripotent and epiblast lineage markers, up-regulation of the Oct4-repressed marker Id2, and unchanged levels of Gapdh and Ahcy. (F_–_I) Alkaline phosphatase staining of mESCs treated with siRNAs against AK028326 (F), AK141205 (G), NS control (H), or mock transfection control (MT control) (I). Scale bar, 100 μm. (Asterisks) Significant difference from control samples (*, P < 0.05; **, P < 0.01; ***, P < 0.001) relative to NS control (dotted red line).
FIGURE 5.
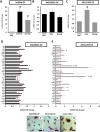
Expression patterns of lncRNAs, pluripotent markers, trophectoderm markers, and epiblast lineage markers in response to lncRNA overexpression (OE). (A) LncRNAs AK028326 and AK141205 are significantly overexpressed relative to the vector-only control. (B) AK028326 overexpression marginally altered Oct4 and Nanog transcription. (C) AK141205 overexpression significantly up-regulated Oct4 but not Nanog transcription relative to vector-only control. (D) In response to AK028326 overexpression, there was significant enhancement of mesodermal marker T/Brachyury and Sox4, the epiblast marker Fgf5, the ectodermal markers Sox11, Pax6, and Pax7, in addition to unchanged levels of the housekeeping genes Gapdh and Ahcy. (E) In response to AK141205 overexpression, there was significant enhancement of mesodermal marker T/Brachyury, the epiblast marker Fgf5, the ectodermal markers Sox1 and Sox11, and endoderm markers, in addition to unchanged levels of Gapdh and Ahcy. (F_–_H) AP staining of AK028326-OE (F), AK141205-OE (G), and vector-only control (H). Scale bar, 100 μm. (Asterisks) Significant difference from control samples (*, P < 0.05; ***, P < 0.001) relative to the pCAG control (dotted red line).
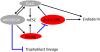
FIGURE 6.
A model for AK028326 and AK141205 lncRNA placement into the Oct4 and Nanog regulatory network. (Black lines) Positive regulation; (blue lines) repression. Oct4 and Nanog are required for self-renewal, and Oct4 activates AK028326, while Nanog represses AK141205. Reduction of AK028326 exhibited a correlated reduction in Oct4 mRNA levels with a concordant up-regulation of Trophoblast lineage transcripts, though the mechanism is unclear. Overexpression of AK141205 led to up-regulation of Oct4 with a corresponding up-regulation of endodermal markers.
Similar articles
- Correlations between the levels of Oct4 and Nanog as a signature for naïve pluripotency in mouse embryonic stem cells.
Muñoz Descalzo S, Rué P, Garcia-Ojalvo J, Martinez Arias A. Muñoz Descalzo S, et al. Stem Cells. 2012 Dec;30(12):2683-91. doi: 10.1002/stem.1230. Stem Cells. 2012. PMID: 22969005 - Gata4 blocks somatic cell reprogramming by directly repressing Nanog.
Serrano F, Calatayud CF, Blazquez M, Torres J, Castell JV, Bort R. Serrano F, et al. Stem Cells. 2013 Jan;31(1):71-82. doi: 10.1002/stem.1272. Stem Cells. 2013. PMID: 23132827 - Simultaneous overexpression of Oct4 and Nanog abrogates terminal myogenesis.
Lang KC, Lin IH, Teng HF, Huang YC, Li CL, Tang KT, Chen SL. Lang KC, et al. Am J Physiol Cell Physiol. 2009 Jul;297(1):C43-54. doi: 10.1152/ajpcell.00468.2008. Epub 2009 Apr 29. Am J Physiol Cell Physiol. 2009. PMID: 19403798 - The transcriptional foundation of pluripotency.
Chambers I, Tomlinson SR. Chambers I, et al. Development. 2009 Jul;136(14):2311-22. doi: 10.1242/dev.024398. Development. 2009. PMID: 19542351 Free PMC article. Review. - Role of Oct4 in maintaining and regaining stem cell pluripotency.
Shi G, Jin Y. Shi G, et al. Stem Cell Res Ther. 2010 Dec 14;1(5):39. doi: 10.1186/scrt39. Stem Cell Res Ther. 2010. PMID: 21156086 Free PMC article. Review.
Cited by
- Activity-dependent human brain coding/noncoding gene regulatory networks.
Lipovich L, Dachet F, Cai J, Bagla S, Balan K, Jia H, Loeb JA. Lipovich L, et al. Genetics. 2012 Nov;192(3):1133-48. doi: 10.1534/genetics.112.145128. Epub 2012 Sep 7. Genetics. 2012. PMID: 22960213 Free PMC article. - Novel Approaches to Profile Functional Long Noncoding RNAs Associated with Stem Cell Pluripotency.
Zhu Y, Yan Z, Tang Z, Li W. Zhu Y, et al. Curr Genomics. 2020 Jan;21(1):37-45. doi: 10.2174/1389202921666200210142840. Curr Genomics. 2020. PMID: 32655297 Free PMC article. Review. - Mutations within lncRNAs are effectively selected against in fruitfly but not in human.
Haerty W, Ponting CP. Haerty W, et al. Genome Biol. 2013 May 27;14(5):R49. doi: 10.1186/gb-2013-14-5-r49. Genome Biol. 2013. PMID: 23710818 Free PMC article. - Regulation of long non-coding RNAs and genome dynamics by the RNA surveillance machinery.
Nair L, Chung H, Basu U. Nair L, et al. Nat Rev Mol Cell Biol. 2020 Mar;21(3):123-136. doi: 10.1038/s41580-019-0209-0. Epub 2020 Feb 4. Nat Rev Mol Cell Biol. 2020. PMID: 32020081 Free PMC article. Review. - Gas5 is an essential lncRNA regulator for self-renewal and pluripotency of mouse embryonic stem cells and induced pluripotent stem cells.
Tu J, Tian G, Cheung HH, Wei W, Lee TL. Tu J, et al. Stem Cell Res Ther. 2018 Mar 21;9(1):71. doi: 10.1186/s13287-018-0813-5. Stem Cell Res Ther. 2018. PMID: 29562912 Free PMC article.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
- Avner P, Heard E. X-chromosome inactivation: Counting, choice, and initiation. Nat Rev Genet. 2001;2:59–67. - PubMed
- Bibel M, Richter J, Schrenk K, Tucker KL, Staiger V, Korte M, Goetz M, Barde YA. Differentiation of mouse embryonic stem cells into a defined neuronal lineage. Nat Neurosci. 2004;7:1003–1009. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials