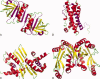Amyloidogenic sequences in native protein structures - PubMed (original) (raw)
Amyloidogenic sequences in native protein structures
Susan Tzotzos et al. Protein Sci. 2010 Feb.
Abstract
Numerous short peptides have been shown to form beta-sheet amyloid aggregates in vitro. Proteins that contain such sequences are likely to be problematic for a cell, due to their potential to aggregate into toxic structures. We investigated the structures of 30 proteins containing 45 sequences known to form amyloid, to see how the proteins cope with the presence of these potentially toxic sequences, studying secondary structure, hydrogen-bonding, solvent accessible surface area and hydrophobicity. We identified two mechanisms by which proteins avoid aggregation: Firstly, amyloidogenic sequences are often found within helices, despite their inherent preference to form beta structure. Helices may offer a selective advantage, since in order to form amyloid the sequence will presumably have to first unfold and then refold into a beta structure. Secondly, amyloidogenic sequences that are found in beta structure are usually buried within the protein. Surface exposed amyloidogenic sequences are not tolerated in strands, presumably because they lead to protein aggregation via assembly of the amyloidogenic regions. The use of alpha-helices, where amyloidogenic sequences are forced into helix, despite their intrinsic preference for beta structure, is thus a widespread mechanism to avoid protein aggregation.
Figures
Figure 1
Selection of amyloidogenic proteins in native conformation with amyloidogenic sequences highlighted (amyloidogenic residue numbers in parentheses for given protein model refer to residues considered amyloidogenic for calculations in present study). Biological molecules are illustrated unless otherwise indicated. a) β-lactoglobulin, PDB ID 1BEB (Asp11-Tyr20, Lys101-Ser110, Ser116-Pro126, His146-Asn152), b) prolactin, PDB ID 1RW5 (Gly7-Ser34, Arg43-Ser57), c) repA pPS10 Pseudomonas, PDB ID 1HKQ (Leu26-Ile34), d) B. subtilis ‘YjcG’ protein, PDB ID 2D4G (Leu151-Asn156). Images were created using PyMol (www.pymol.org).
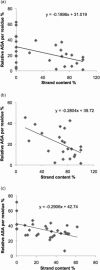
Figure 2
(a) Correlation between relative accessible surface area (ASA) per residue and strand content in amyloidogenic sequences of 30 native proteins (one average value per protein). Correlation coefficient = −0.4568, p = 0.0112; slope of trendline = −0.1896. (b) Correlation between relative accessible surface area (ASA) per residue and strand content in S-set of amyloidogenic sequences. Correlation coefficient = −0.5239, p = 0.0123; slope of the trendline = −0.2804. (c) Correlation between relative accessible surface area (ASA) per residue and strand content in non-amyloidogenic sequences of 30 native proteins. Correlation coefficient = −0.4735, p = 0.0082; slope of the trendline = −0.2906.
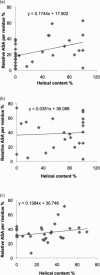
Figure 3
(a) Correlation between relative accessible surface area (ASA) per residue and helical content in amyloidogenic sequences of 30 native proteins (one average value per protein). Correlation coefficient = 0.4244, p = 0.0194; slope of trendline = 0.1744. (b) Correlation between relative accessible surface area (ASA) per residue and helical content in H-set of amyloidogenic sequences. Correlation coefficient = 0.0610, p = 0.7823; slope of the trendline = −0.0351. (c) Correlation between relative accessible surface area (ASA) per residue and helical content in non-amyloidogenic sequences of 30 native proteins. Correlation coefficient = 0.2414, p = 0.1987; slope of the trendline = 0.1384.
Similar articles
- Structure-based design and study of non-amyloidogenic, double N-methylated IAPP amyloid core sequences as inhibitors of IAPP amyloid formation and cytotoxicity.
Kapurniotu A, Schmauder A, Tenidis K. Kapurniotu A, et al. J Mol Biol. 2002 Jan 18;315(3):339-50. doi: 10.1006/jmbi.2001.5244. J Mol Biol. 2002. PMID: 11786016 - Molecular mechanism of β-sheet self-organization at water-hydrophobic interfaces.
Nikolic A, Baud S, Rauscher S, Pomès R. Nikolic A, et al. Proteins. 2011 Jan;79(1):1-22. doi: 10.1002/prot.22854. Epub 2010 Oct 11. Proteins. 2011. PMID: 20938982 - Covalently attached fatty acyl chains alter the aggregation behavior of an amyloidogenic peptide derived from human β(2)-microglobulin.
Rawat A, Nagaraj R. Rawat A, et al. J Pept Sci. 2013 Dec;19(12):770-83. doi: 10.1002/psc.2575. Epub 2013 Oct 31. J Pept Sci. 2013. PMID: 24243599 - Amyloid peptides and proteins in review.
Harrison RS, Sharpe PC, Singh Y, Fairlie DP. Harrison RS, et al. Rev Physiol Biochem Pharmacol. 2007;159:1-77. doi: 10.1007/112_2007_0701. Rev Physiol Biochem Pharmacol. 2007. PMID: 17846922 Review. - Molecular mechanisms of polypeptide aggregation in human diseases.
Khare SD, Dokholyan NV. Khare SD, et al. Curr Protein Pept Sci. 2007 Dec;8(6):573-9. doi: 10.2174/138920307783018703. Curr Protein Pept Sci. 2007. PMID: 18220844 Review.
Cited by
- Peptide-Induced Amyloid-Like Conformational Transitions in Proteins.
Egorov V, Grudinina N, Vasin A, Lebedev D. Egorov V, et al. Int J Pept. 2015;2015:723186. doi: 10.1155/2015/723186. Epub 2015 Sep 8. Int J Pept. 2015. PMID: 26435719 Free PMC article. Review. - X-ray Crystallographic Structures of Oligomers of Peptides Derived from β2-Microglobulin.
Spencer RK, Kreutzer AG, Salveson PJ, Li H, Nowick JS. Spencer RK, et al. J Am Chem Soc. 2015 May 20;137(19):6304-11. doi: 10.1021/jacs.5b01673. Epub 2015 May 12. J Am Chem Soc. 2015. PMID: 25915729 Free PMC article. - The N-terminal helix controls the transition between the soluble and amyloid states of an FF domain.
Castillo V, Chiti F, Ventura S. Castillo V, et al. PLoS One. 2013;8(3):e58297. doi: 10.1371/journal.pone.0058297. Epub 2013 Mar 7. PLoS One. 2013. PMID: 23505482 Free PMC article. - The proteasome as a druggable target with multiple therapeutic potentialities: Cutting and non-cutting edges.
Tundo GR, Sbardella D, Santoro AM, Coletta A, Oddone F, Grasso G, Milardi D, Lacal PM, Marini S, Purrello R, Graziani G, Coletta M. Tundo GR, et al. Pharmacol Ther. 2020 Sep;213:107579. doi: 10.1016/j.pharmthera.2020.107579. Epub 2020 May 19. Pharmacol Ther. 2020. PMID: 32442437 Free PMC article. - Association between foldability and aggregation propensity in small disulfide-rich proteins.
Fraga H, Graña-Montes R, Illa R, Covaleda G, Ventura S. Fraga H, et al. Antioxid Redox Signal. 2014 Jul 20;21(3):368-83. doi: 10.1089/ars.2013.5543. Epub 2014 May 5. Antioxid Redox Signal. 2014. PMID: 24635049 Free PMC article.
References
- Dobson CM. Protein folding and misfolding. Nature. 2003;426:884–890. - PubMed
- Thirumalai D, Klimov DK, Dima RI. Emerging ideas on the molecular basis of protein and peptide aggregation. Curr Opin Struct Biol. 2003;13:146–159. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources