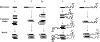Beta arcades: recurring motifs in naturally occurring and disease-related amyloid fibrils - PubMed (original) (raw)
Review
Beta arcades: recurring motifs in naturally occurring and disease-related amyloid fibrils
Andrey V Kajava et al. FASEB J. 2010 May.
Abstract
Amyloid fibrils are filamentous protein aggregates that accumulate in diseases such as Alzheimer's or type II diabetes. The amyloid-forming protein is disease specific. Amyloids may also be formed in vitro from many other proteins, after first denaturing them. Unlike the diverse native folds of these proteins, their amyloids are fundamentally similar in being rigid, smooth-sided, and cross-beta-structured, that is, with beta strands running perpendicular to the fibril axis. In the absence of high-resolution fibril structures, increasingly credible models are being derived by integrating data from a crossfire of experimental techniques. Most current models of disease-related amyloids invoke "beta arcades," columnar structures produced by in-register stacking of "beta arches." A beta arch is a strand-turn-strand motif in which the two beta strands interact via their side chains, not via the polypeptide backbone as in a conventional beta hairpin. Crystal structures of beta-solenoids, a class of proteins with amyloid-like properties, offer insight into the beta-arc turns found in beta arches. General conformational and thermodynamic considerations suggest that complexes of 2 or more beta arches may nucleate amyloid fibrillogenesis in vivo. The apparent prevalence of beta arches and their components have implications for identifying amyloidogenic sequences, elucidating fibril polymorphisms, predicting the locations and conformations of beta arcs within amyloid fibrils, and refining existing fibril models.
Figures
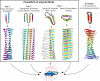
Figure 1.
Three types of cross-β models for amyloid protofibrils. Top: axial views of the repetitive structural units (rectangles represent β strands); bottom: lateral views of protofibrils formed by stacking the corresponding repetitive units. Orange circles in the insulin model show Cys residues forming disulfide bonds. Beneath, schematic diagram of a β arcade, considered as a structural motif common to all 3 types of models. One β arch is colored in blue, with depth cuing; arrows indicate β strands; dotted lines show H bonds. Right: crystal structure of the SufD dimer , shown as an illustrative β-solenoid with a stacking of β arches similar to that envisaged in the amyloid models.
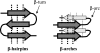
Figure 2.
β hairpins and β arches. Arrows indicate β strands, dotted lines show H bonds.
Figure 3.
Lateral and axial views of some structures containing inter-β-arc H bonding. Left: crystal structure of yeast CAP protein ; middle: crystal structure of antifreeze protein from Tenebrio molitor ; right: model of corrugated β-structure of Aβ (unpublished results) built with reference to cryo-electron microscopy data . Only the polypeptide backbone is shown (except in the axial view of the corrugated structure). Dotted lines show H bonds.
Figure 4.
Hypothetical pathways of amyloid fibril assembly. Bars represent β strands; curved lines represent other parts of the polypeptide chains. In some cases, two similar but nonidentical strand-forming peptides are shown in different shades. Thin lines between bars represent H bonds. Pathway 1: short amyloid-forming peptides (<15 residues or so) form dimers, trimers (transition state), and then tetrameric double-β-sheet nucleus . Pathway 2: long peptides (>15 residues or so) form a β-arch (transition state) and then a double-β-sheet nucleus. Pathway 2a: long peptides (>15 residues or so) form β hairpins, and then 2 β hairpins side by side (transition state), which transforms into a double-β-sheet nucleus. Pathway 3: short amyloid-forming peptides with nonamyloidogenic flanking regions follow a pathway similar to pathway 1. Pathway 4: long peptides with nonamyloidogenic flanking regions follow a pathway similar to those of pathway 2 or 2a_._
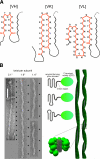
Figure 5.
Distinct cross-β structures. Left: a fibril formed from antiparallel cross β-structures, in this case β meanders packed side by side; middle: fibril formed by a stacking of β-solenoids (one shown with black turns; one with blue turns); right: fibril formed from in-register stacking of β arches (parallel superpleated structure). In these examples, each monomer has 4 β strands (arrows). Gln side chains in H-bonded ladders located inside the fibrils are shown in red. Bottom: red circles indicate locations of Gln residues (Q) within the corresponding peptides that form H-bonded ladders. Models are arranged from left to right in order of progressive relaxation of the requirement for specific sequence motifs. In the model at right, a Gln in any position produces a ladder. Ladders based on differing serpentine folds of the same polypeptide chain are, in principle, possible, which would provide a structural basis for fibril polymorphism. The other two structures require a specific distance between the Glns for a ladder to be formed, and this constraint reduces the number of possible structures. We consider that antiparallel β-structured fibrils formed from peptides that are rich in Gln and/or Asn and whose stability depends on ladders of these residues are likely to be rare.
Figure 6.
A) Polymorphism of Sup35p fibrils. Cross-sections of hypothetical models of Sup35 related to the strains of the yeast prion [PSI]: VH strain requires residues 7–21; VK strain requires residues 9–37; VL strain requires residues 5–52 . Each model envisages a parallel in-register stacking of the corresponding serpentine. Primary β arches are outlined in red; secondary pleats are in black. B) Axial repeat length polymorphism in Ure2p-derived filaments. Left: electron micrographs of negatively stained specimens of Ure2p (1–80)-GFP filaments that exhibit different axial repeat lengths . Repeats are indexed with black arrowheads at right of each filament. Scale bar = 100 nm. Right: schematic representation of 3 different variants that lead to different repeat lengths. We hypothesize that this polymorphism arises from different serpentine folds of the prion domain that result in slightly differing axial twists per subunit when the subunits are stacked in parallel in-register arrangements. Specifics of the envisaged folds are unknown, and the examples shown are simply for illustrative purposes. The corkscrew appearance of the filaments arises from clusters of globular GFP moieties precessing around the amyloid fibril backbone (middle, bottom).
Similar articles
- Standard conformations of beta-arches in beta-solenoid proteins.
Hennetin J, Jullian B, Steven AC, Kajava AV. Hennetin J, et al. J Mol Biol. 2006 May 12;358(4):1094-105. doi: 10.1016/j.jmb.2006.02.039. Epub 2006 Mar 2. J Mol Biol. 2006. PMID: 16580019 - Elucidating the Structures of Amyloid Oligomers with Macrocyclic β-Hairpin Peptides: Insights into Alzheimer's Disease and Other Amyloid Diseases.
Kreutzer AG, Nowick JS. Kreutzer AG, et al. Acc Chem Res. 2018 Mar 20;51(3):706-718. doi: 10.1021/acs.accounts.7b00554. Epub 2018 Mar 6. Acc Chem Res. 2018. PMID: 29508987 Free PMC article. Review. - Site-specific identification of non-beta-strand conformations in Alzheimer's beta-amyloid fibrils by solid-state NMR.
Antzutkin ON, Balbach JJ, Tycko R. Antzutkin ON, et al. Biophys J. 2003 May;84(5):3326-35. doi: 10.1016/S0006-3495(03)70057-5. Biophys J. 2003. PMID: 12719262 Free PMC article. - Synchrotron X-ray studies suggest that the core of the transthyretin amyloid fibril is a continuous beta-sheet helix.
Blake C, Serpell L. Blake C, et al. Structure. 1996 Aug 15;4(8):989-98. doi: 10.1016/s0969-2126(96)00104-9. Structure. 1996. PMID: 8805583 - A brief overview of amyloids and Alzheimer's disease.
Ow SY, Dunstan DE. Ow SY, et al. Protein Sci. 2014 Oct;23(10):1315-31. doi: 10.1002/pro.2524. Epub 2014 Jul 30. Protein Sci. 2014. PMID: 25042050 Free PMC article. Review.
Cited by
- Intrinsically disordered proteins of viruses: Involvement in the mechanism of cell regulation and pathogenesis.
Mishra PM, Verma NC, Rao C, Uversky VN, Nandi CK. Mishra PM, et al. Prog Mol Biol Transl Sci. 2020;174:1-78. doi: 10.1016/bs.pmbts.2020.03.001. Epub 2020 Apr 2. Prog Mol Biol Transl Sci. 2020. PMID: 32828463 Free PMC article. Review. - Insights into protein misfolding and aggregation enabled by solid-state NMR spectroscopy.
van der Wel PCA. van der Wel PCA. Solid State Nucl Magn Reson. 2017 Nov;88:1-14. doi: 10.1016/j.ssnmr.2017.10.001. Epub 2017 Oct 4. Solid State Nucl Magn Reson. 2017. PMID: 29035839 Free PMC article. Review. - The amyloid structure of mouse RIPK3 (receptor interacting protein kinase 3) in cell necroptosis.
Wu XL, Hu H, Dong XQ, Zhang J, Wang J, Schwieters CD, Liu J, Wu GX, Li B, Lin JY, Wang HY, Lu JX. Wu XL, et al. Nat Commun. 2021 Mar 12;12(1):1627. doi: 10.1038/s41467-021-21881-2. Nat Commun. 2021. PMID: 33712586 Free PMC article. - Protein-conformational diseases in childhood: Naturally-occurring hIAPP amyloid-oligomers and early β-cell damage in obesity and diabetes.
Altamirano-Bustamante NF, Garrido-Magaña E, Morán E, Calderón A, Pasten-Hidalgo K, Castillo-Rodríguez RA, Rojas G, Lara-Martínez R, Leyva-García E, Larralde-Laborde M, Domíguez G, Murata C, Margarita-Vazquez Y, Payro R, Barbosa M, Valderrama A, Montesinos H, Domínguez-Camacho A, García-Olmos VH, Ferrer R, Medina-Bravo PG, Santoscoy F, Revilla-Monsalve C, Jiménez-García LF, Morán J, Villalobos-Alva J, Villalobos MJ, Calzada-León R, Altamirano P, Altamirano-Bustamante MM. Altamirano-Bustamante NF, et al. PLoS One. 2020 Aug 24;15(8):e0237667. doi: 10.1371/journal.pone.0237667. eCollection 2020. PLoS One. 2020. PMID: 32833960 Free PMC article. - W8, a new Sup35 prion strain, transmits distinctive information with a conserved assembly scheme.
Huang YW, Chang YC, Diaz-Avalos R, King CY. Huang YW, et al. Prion. 2015;9(3):207-27. doi: 10.1080/19336896.2015.1039217. Prion. 2015. PMID: 26038983 Free PMC article.
References
- Pepys M B. Amyloidosis. Annu Rev Med. 2006;57:223–241. - PubMed
- Stefani M, Dobson C M. Protein aggregation and aggregate toxicity: new insights into protein folding, misfolding diseases and biological evolution. J Mol Med. 2003;81:678–699. - PubMed
- Lashuel H A, Lansbury P T., Jr Are amyloid diseases caused by protein aggregates that mimic bacterial pore-forming toxins? Q Rev Biophys. 2006;39:167–201. - PubMed
- King C Y, Diaz-Avalos R. Protein-only transmission of three yeast prion strains. Nature. 2004;428:319–323. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical