Novel mutant-selective EGFR kinase inhibitors against EGFR T790M - PubMed (original) (raw)
. 2009 Dec 24;462(7276):1070-4.
doi: 10.1038/nature08622.
Dalia Ercan, Liang Chen, Cai-Hong Yun, Danan Li, Marzia Capelletti, Alexis B Cortot, Lucian Chirieac, Roxana E Iacob, Robert Padera, John R Engen, Kwok-Kin Wong, Michael J Eck, Nathanael S Gray, Pasi A Jänne
Affiliations
- PMID: 20033049
- PMCID: PMC2879581
- DOI: 10.1038/nature08622
Novel mutant-selective EGFR kinase inhibitors against EGFR T790M
Wenjun Zhou et al. Nature. 2009.
Abstract
The clinical efficacy of epidermal growth factor receptor (EGFR) kinase inhibitors in EGFR-mutant non-small-cell lung cancer (NSCLC) is limited by the development of drug-resistance mutations, including the gatekeeper T790M mutation. Strategies targeting EGFR T790M with irreversible inhibitors have had limited success and are associated with toxicity due to concurrent inhibition of wild-type EGFR. All current EGFR inhibitors possess a structurally related quinazoline-based core scaffold and were identified as ATP-competitive inhibitors of wild-type EGFR. Here we identify a covalent pyrimidine EGFR inhibitor by screening an irreversible kinase inhibitor library specifically against EGFR T790M. These agents are 30- to 100-fold more potent against EGFR T790M, and up to 100-fold less potent against wild-type EGFR, than quinazoline-based EGFR inhibitors in vitro. They are also effective in murine models of lung cancer driven by EGFR T790M. Co-crystallization studies reveal a structural basis for the increased potency and mutant selectivity of these agents. These mutant-selective irreversible EGFR kinase inhibitors may be clinically more effective and better tolerated than quinazoline-based inhibitors. Our findings demonstrate that functional pharmacological screens against clinically important mutant kinases represent a powerful strategy to identify new classes of mutant-selective kinase inhibitors.
Figures
Figure 1. WZ4002, WZ3146 and WZ8040 are novel EGFR inhibitors, suppress the growth of EGFR T790M containing cell lines and inhibit EGFR phosphorylation
A. Chemical structures of the WZ compounds. B. IC50 values (nM) for NSCLC cell lines (top) and Ba/F3 (bottom) cells, with genotypes corresponding to the NSCLC cell lines, treated with indicated drugs. Growth was assessed using the MTS survival assay. C. Comparison of WZ3146, WZ4002 and CL-387,785 on EGFR signaling in PC9 GR cells. The cells were treated with the indicated concentrations of each drug for 16 hours. Cell extracts were immunoblotted to detect the indicated proteins. D. Comparison of EGFR inhibitors on EGFR phosphorylation in 3T3 cells expressing del E746_A750/T90M. The cells were treated with indicated concentrations of each drug for 16 hours and stimulated with EGF (10 ng/ml) 15 minutes prior to lysis. Cell extracts were immunoblotted to detect the indicated proteins.
Figure 2. WZ4002 is less potent than quinazoline EGFR inhibitors against wild type EGFR in vitro and in vivo
A. EGFR vIII Ba/F3 cells treated with WZ or quinazoline EGFR inhibitors. The mean (n=6) and standard deviation is plotted for each drug and concentration. B. Comparison of EGFR inhibitors on EGFR phosphorylation in 3T3 cells expressing wild type EGFR. The cells were treated with indicated concentrations of each drug for 16 hours and stimulated with EGF (10 ng/ml) 15 minutes prior to lysis. Cell extracts were immunoblotted to detect the indicated proteins. C. Immunohistochemical analysis of skin from erlotinib or WZ4002 treated mice using EGFR and pY1173 EGFR. Only erlotinib treatment results in significant inhibition of EGFR phosphorylation. Scale bar, 50 μm. D. Quantification of frequency of phospho-EGFR staining from vehicle (n=3), erlotinib (n=3) and WZ4002 (n=2) treated mice. The mean and standard deviations are plotted for drug treatment.
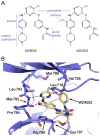
Figure 3. Crystal Structure of WZ4002 bound to EGFR T790M
A. Chemical structures of WZ8040 and WZ4002 are shown schematically in a manner resembling the conformation adopted in complex with the kinase. B. Crystal structure of WZ4002 in complex with EGFR T790M mutant (PDB ID 3IKA). WZ4002 binds the active conformation of the kinase, with the both the regulatory C-helix and “DFG” segment of the activation loop in their inward, active positions. The EGFR kinase is shown in a ribbon representation (blue) with the bound inhibitor in yellow. Sidechain and mainchain atoms are shown for selected residues that contact the compound. Expected hydrogen bonds to the backbone amide and carbonyl atoms of Met 793 are indicated by dashed lines. Note also the covalent bond with Cys797. The structure was refined to a crystallographic R value of 21.3% (Rfree=25.4%) with data extending to 2.9Å resolution (see methods for further crystallographic details).
Figure 4. WZ4002 inhibits EGFR phosphorylation and induces significant tumor regression in murine models of EGFR T790M
A. Two doses separated by 16 hours of WZ4002 (2.5 mg/kg or 25 mg/kg) or vehicle were administered to EGFR delE746_A750/T790M or L858R/T790M mice with MRI confirmed tumors. The mice were sacrificed, the lungs isolated and grossly dissected and subjected to cell lysis. Cell extracts were immunoblotted to detect the indicated proteins. B. Immunohistochemical analyses of tumors from EGFR delE746_A750/T790M mice from A. using indicated antibodies. Scale bar, 50 μm. C. Quantification of TUNEL and Ki67 positive cells from tumor nodules (n=4) from vehicle and WZ4002 treated mice. The mean and standard deviation are plotted. D. MRI images of vehicle or WZ4002 treated mice at baseline (0w) and following 2 weeks (2w) of treatment. E. Quantification of the relative tumor volume from MRI images from vehicle treated mice (E746_A750/T790M (n=3); L858R/T790M (n = 4)), and WZ4002 treated L858R/T790M (n=3) and E746_A750/T790M (n=3) mice. The mean and standard deviation are plotted. F. Tumors from vehicle and WZ4002 treated mice stained with hematoxylin and eosin. Low power view (inset) demonstrates near complete resolution of tumors in the WZ4002 treated mice. Scale bar, 100 μm.
Similar articles
- Overcoming EGFR(T790M) and EGFR(C797S) resistance with mutant-selective allosteric inhibitors.
Jia Y, Yun CH, Park E, Ercan D, Manuia M, Juarez J, Xu C, Rhee K, Chen T, Zhang H, Palakurthi S, Jang J, Lelais G, DiDonato M, Bursulaya B, Michellys PY, Epple R, Marsilje TH, McNeill M, Lu W, Harris J, Bender S, Wong KK, Jänne PA, Eck MJ. Jia Y, et al. Nature. 2016 Jun 2;534(7605):129-32. doi: 10.1038/nature17960. Epub 2016 May 25. Nature. 2016. PMID: 27251290 Free PMC article. - Discovery of selective irreversible inhibitors for EGFR-T790M.
Zhou W, Ercan D, Jänne PA, Gray NS. Zhou W, et al. Bioorg Med Chem Lett. 2011 Jan 15;21(2):638-43. doi: 10.1016/j.bmcl.2010.12.036. Epub 2010 Dec 10. Bioorg Med Chem Lett. 2011. PMID: 21208802 Free PMC article. - Novel Selective and Potent EGFR Inhibitor that Overcomes T790M-Mediated Resistance in Non-Small Cell Lung Cancer.
Li Y, Song Z, Jin Y, Tang Z, Kang J, Ma X. Li Y, et al. Molecules. 2016 Nov 2;21(11):1462. doi: 10.3390/molecules21111462. Molecules. 2016. PMID: 27827863 Free PMC article. - Fighting cancer drug resistance: Opportunities and challenges for mutation-specific EGFR inhibitors.
Juchum M, Günther M, Laufer SA. Juchum M, et al. Drug Resist Updat. 2015 May;20:12-28. doi: 10.1016/j.drup.2015.05.002. Epub 2015 May 12. Drug Resist Updat. 2015. PMID: 26021435 Review. - Targeting EGFRL858R/T790M and EGFRL858R/T790M/C797S resistance mutations in NSCLC: Current developments in medicinal chemistry.
Lu X, Yu L, Zhang Z, Ren X, Smaill JB, Ding K. Lu X, et al. Med Res Rev. 2018 Sep;38(5):1550-1581. doi: 10.1002/med.21488. Epub 2018 Jan 26. Med Res Rev. 2018. PMID: 29377179 Review.
Cited by
- Discovery of a Pyrrolopyrimidine (JH-II-127), a Highly Potent, Selective, and Brain Penetrant LRRK2 Inhibitor.
Hatcher JM, Zhang J, Choi HG, Ito G, Alessi DR, Gray NS. Hatcher JM, et al. ACS Med Chem Lett. 2015 Apr 7;6(5):584-9. doi: 10.1021/acsmedchemlett.5b00064. eCollection 2015 May 14. ACS Med Chem Lett. 2015. PMID: 26005538 Free PMC article. - In vivo PET Imaging of EGFR Expression: An Overview of Radiolabeled EGFR TKIs.
Zhu J, Li Y, Wu X, Li Y, Wang L, Fan H. Zhu J, et al. Curr Top Med Chem. 2022;22(28):2329-2342. doi: 10.2174/1568026622666220903142416. Curr Top Med Chem. 2022. PMID: 36056825 - A Radiobrominated Tyrosine Kinase Inhibitor for EGFR with L858R/T790M Mutations in Lung Carcinoma.
Fawwaz M, Mishiro K, Nishii R, Makino A, Kiyono Y, Shiba K, Kinuya S, Ogawa K. Fawwaz M, et al. Pharmaceuticals (Basel). 2021 Mar 12;14(3):256. doi: 10.3390/ph14030256. Pharmaceuticals (Basel). 2021. PMID: 33809064 Free PMC article. - Emerging Agents and New Mutations in EGFR-Mutant Lung Cancer.
Ayeni D, Politi K, Goldberg SB. Ayeni D, et al. Clin Cancer Res. 2015 Sep 1;21(17):3818-20. doi: 10.1158/1078-0432.CCR-15-1211. Epub 2015 Jul 13. Clin Cancer Res. 2015. PMID: 26169963 Free PMC article. - Mechanism of Resistance to Epidermal Growth Factor Receptor-Tyrosine Kinase Inhibitors and a Potential Treatment Strategy.
Nagano T, Tachihara M, Nishimura Y. Nagano T, et al. Cells. 2018 Nov 15;7(11):212. doi: 10.3390/cells7110212. Cells. 2018. PMID: 30445769 Free PMC article. Review.
References
- Mok TS, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. - PubMed
- Kobayashi S, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–792. - PubMed
- Li D, et al. Bronchial and peripheral murine lung carcinomas induced by T790M-L858R mutant EGFR respond to HKI-272 and rapamycin combination therapy. Cancer Cell. 2007;12:81–93. - PubMed
- Besse B, et al. Neratinib (HKI-272), an irreversible pan-ErbB receptor tyrosine kinase inhibtor: preliminary results of a phase 2 tiral in patients with advanced non-small cell lung cancer. European Journal of Cancer. 2008;6:64. abstract 203.
Publication types
MeSH terms
Substances
Grants and funding
- P50CA090578/CA/NCI NIH HHS/United States
- R01CA116020/CA/NCI NIH HHS/United States
- R01 GM070590/GM/NIGMS NIH HHS/United States
- R01GM070590/GM/NIGMS NIH HHS/United States
- R01 CA080942/CA/NCI NIH HHS/United States
- R01CA080942/CA/NCI NIH HHS/United States
- R01 CA135257/CA/NCI NIH HHS/United States
- R01 CA130876-02/CA/NCI NIH HHS/United States
- R01 CA130876/CA/NCI NIH HHS/United States
- R01 CA122794/CA/NCI NIH HHS/United States
- R01AG2400401/AG/NIA NIH HHS/United States
- R01CA11446/CA/NCI NIH HHS/United States
- R01CA135257/CA/NCI NIH HHS/United States
- R01CA130876-02/CA/NCI NIH HHS/United States
- P50 CA090578/CA/NCI NIH HHS/United States
- R01 CA116020/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous