Mammalian autophagy: core molecular machinery and signaling regulation - PubMed (original) (raw)
Review
Mammalian autophagy: core molecular machinery and signaling regulation
Zhifen Yang et al. Curr Opin Cell Biol. 2010 Apr.
Abstract
Autophagy, a cellular catabolic pathway, is evolutionarily conserved from yeast to mammals. Central to this process is the formation of autophagosomes, double-membrane vesicles responsible for delivering long-lived proteins and excess or damaged organelle into the lysosome for degradation and reuse of the resulting macromolecules. In addition to the hallmark discovery of core molecular machinery components involved in autophagosome formation, complex signaling cascades controlling autophagy have also begun to emerge, with mTOR as a central but far from exclusive player. Malfunction of autophagy has been linked to a wide range of human pathologies, including cancer, neurodegeneration, and pathogen infection. Here we highlight the recent advances in identifying and understanding the core molecular machinery and signaling pathways that are involved in mammalian autophagy.
Keywords: autophagy; lysosomes; mammalian cells; signal transduction; stress.
Copyright 2009 Elsevier Ltd. All rights reserved.
Figures
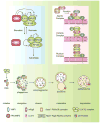
Figure 1. Schematic depiction of the autophagy pathway and its core molecular machinery in mammalian cells
Mammalian autophagy proceeds through a series of steps, including initiation at the PAS (phagophore assembly site), elongation and expansion of the phagophore, closure and completion of the autophagosome, autophagosome maturation via docking and fusion with an endosome and/or lysosome, breakdown and degradation of the autophagosome inner membrane and cargo, and recycling of the resulting macromolecules. Regulatory components for autophagy induction include the ULK1 and ULK2 complexes that contain various Atg proteins (light blue box at left) that are required for autophagy. The association of mTORC1 with this complex and the activity of mTORC1 depend on the nutrient status. Under nutrient-rich conditions, mTORC1 is associated with the ULK1 and ULK2 complexes, and phosphorylates ULK1, ULK2, and mAtg13; upon inactivation of mTORC1 by nutrient starvation, mTORC1 disassociates, mAtg13, ULK1 and ULK2 are partially dephosphorylated, and activation of ULK1 and ULK2 promotes phosphosphorylation of FIP200. There are at least three class III PtdIns3K complexes (light red box at right), that are involved in autophagosome formation or clearance. The Atg14L (Atg14L-Beclin 1-hVps34-p150) and UVRAG (UVRAG-Beclin 1-hVps34-p150) complexes are required for autophagy, whereas the Rubicon complex (Rubicon-UVRAG-Beclin 1-hVps34-p150) negatively regulates autophagy. Ambra1 and Bif-1 are essential for induction of autophagy, through direct interaction with Beclin 1 and UVRAG, respectively, whereas Bcl-2 binds to Beclin 1 and disrupts the Beclin 1-associated hVps34 complex, thereby inhibiting autophagy.
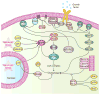
Figure 2. Signaling cascades involved in the regulation of mammalian autophagy
Autophagy is regulated by a complex signaling network of various stimulatory (arrowheads) and inhibitory (bars) inputs. Activation of growth factor receptors stimulates the class I PtdIns3K complex and small GTPase Ras, which leads to activation of the PtdIns3K-PKB-mTORC1 pathway and the Raf-1-MEK1/2-ERK1/2 pathway, respectively. PKB and ERK1/2 phosphorylate and inhibit the GTPase-activating protein complex TSC1/TSC2, leading to the stabilization of Rheb-GTPase, which, in turn, activates mTORC1, causing inhibition of autophagy. Activated ERK1/2 also stimulates autophagy. mTORC2 inhibits autophagy through the phosphorylation and activation of PKB. Metabolic stress, such as high AMP/ATP ratios resulting from energy depletion, or an increase in the cytosolic free Ca2+ concentration or cytokines, cause the AMP-activated protein kinase (AMPK) to be phosphorylated and activated by LKB1, CaMKKβ and TAK1, respectively. AMPK phosphorylates and activates TSC1/TSC2, leading to inactivation of mTORC1 and autophagy induction. Genotoxic and oncogenic stresses result in nuclear p53 stabilization and activation, which stimulates autophagy through activation of AMPK or upregulation of DRAM. In contrast, cytosolic p53 has an inhibitory effect on autophagy. Anti-apoptotic proteins, Bcl-2 or Bcl-XL, associate with Beclin 1 and inhibit the Beclin 1-associated class III PtdIns3K complex, causing inhibition of autophagy. For additional details, see the main text.
Similar articles
- Regulation of mammalian autophagy in physiology and pathophysiology.
Ravikumar B, Sarkar S, Davies JE, Futter M, Garcia-Arencibia M, Green-Thompson ZW, Jimenez-Sanchez M, Korolchuk VI, Lichtenberg M, Luo S, Massey DC, Menzies FM, Moreau K, Narayanan U, Renna M, Siddiqi FH, Underwood BR, Winslow AR, Rubinsztein DC. Ravikumar B, et al. Physiol Rev. 2010 Oct;90(4):1383-435. doi: 10.1152/physrev.00030.2009. Physiol Rev. 2010. PMID: 20959619 Review. - Autophagosome formation: core machinery and adaptations.
Xie Z, Klionsky DJ. Xie Z, et al. Nat Cell Biol. 2007 Oct;9(10):1102-9. doi: 10.1038/ncb1007-1102. Nat Cell Biol. 2007. PMID: 17909521 Review. - Apoptosis and autophagy: Targeting autophagy signalling in cancer cells -'trick or treats'?
Corcelle EA, Puustinen P, Jäättelä M. Corcelle EA, et al. FEBS J. 2009 Nov;276(21):6084-96. doi: 10.1111/j.1742-4658.2009.07332.x. Epub 2009 Sep 29. FEBS J. 2009. PMID: 19788415 Review. - Regulation mechanisms and signaling pathways of autophagy.
He C, Klionsky DJ. He C, et al. Annu Rev Genet. 2009;43:67-93. doi: 10.1146/annurev-genet-102808-114910. Annu Rev Genet. 2009. PMID: 19653858 Free PMC article. Review. - From signal transduction to autophagy of plant cell organelles: lessons from yeast and mammals and plant-specific features.
Reumann S, Voitsekhovskaja O, Lillo C. Reumann S, et al. Protoplasma. 2010 Dec;247(3-4):233-56. doi: 10.1007/s00709-010-0190-0. Epub 2010 Aug 24. Protoplasma. 2010. PMID: 20734094 Review.
Cited by
- A proteomics view of the molecular mechanisms and biomarkers of glaucomatous neurodegeneration.
Tezel G. Tezel G. Prog Retin Eye Res. 2013 Jul;35:18-43. doi: 10.1016/j.preteyeres.2013.01.004. Epub 2013 Feb 5. Prog Retin Eye Res. 2013. PMID: 23396249 Free PMC article. Review. - A novel ER-localized transmembrane protein, EMC6, interacts with RAB5A and regulates cell autophagy.
Li Y, Zhao Y, Hu J, Xiao J, Qu L, Wang Z, Ma D, Chen Y. Li Y, et al. Autophagy. 2013 Feb 1;9(2):150-63. doi: 10.4161/auto.22742. Epub 2012 Nov 26. Autophagy. 2013. PMID: 23182941 Free PMC article. - Protective role of autophagy and autophagy-related protein 5 in early tumorigenesis.
Liu H, He Z, Simon HU. Liu H, et al. J Mol Med (Berl). 2015 Feb;93(2):159-64. doi: 10.1007/s00109-014-1241-3. Epub 2014 Dec 23. J Mol Med (Berl). 2015. PMID: 25529049 Review. - Autophagy and its role in regeneration and remodeling within invertebrate.
Song Q, Liu H, Zhen H, Zhao B. Song Q, et al. Cell Biosci. 2020 Sep 21;10:111. doi: 10.1186/s13578-020-00467-3. eCollection 2020. Cell Biosci. 2020. PMID: 32974004 Free PMC article. Review. - A functional outside-in signaling network of proteoglycans and matrix molecules regulating autophagy.
Neill T, Kapoor A, Xie C, Buraschi S, Iozzo RV. Neill T, et al. Matrix Biol. 2021 Jun;100-101:118-149. doi: 10.1016/j.matbio.2021.04.001. Epub 2021 Apr 7. Matrix Biol. 2021. PMID: 33838253 Free PMC article.
References
- Massey AC, Zhang C, Cuervo AM. Chaperone-mediated autophagy in aging and disease. Curr Top Dev Biol. 2006;73:205–235. - PubMed
- Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous