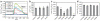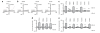Alterations in the ankyrin domain of TRPV4 cause congenital distal SMA, scapuloperoneal SMA and HMSN2C - PubMed (original) (raw)
doi: 10.1038/ng.508. Epub 2009 Dec 27.
Andrea Olschewski, Lea Papić, Hannie Kremer, Meriel E McEntagart, Sabine Uhrig, Carina Fischer, Eleonore Fröhlich, Zoltán Bálint, Bi Tang, Heimo Strohmaier, Hanns Lochmüller, Beate Schlotter-Weigel, Jan Senderek, Angelika Krebs, Katherine J Dick, Richard Petty, Cheryl Longman, Neil E Anderson, George W Padberg, Helenius J Schelhaas, Conny M A van Ravenswaaij-Arts, Thomas R Pieber, Andrew H Crosby, Christian Guelly
Affiliations
- PMID: 20037588
- PMCID: PMC3272392
- DOI: 10.1038/ng.508
Alterations in the ankyrin domain of TRPV4 cause congenital distal SMA, scapuloperoneal SMA and HMSN2C
Michaela Auer-Grumbach et al. Nat Genet. 2010 Feb.
Abstract
Spinal muscular atrophies (SMA, also known as hereditary motor neuropathies) and hereditary motor and sensory neuropathies (HMSN) are clinically and genetically heterogeneous disorders of the peripheral nervous system. Here we report that mutations in the TRPV4 gene cause congenital distal SMA, scapuloperoneal SMA, HMSN 2C. We identified three missense substitutions (R269H, R315W and R316C) affecting the intracellular N-terminal ankyrin domain of the TRPV4 ion channel in five families. Expression of mutant TRPV4 constructs in cells from the HeLa line revealed diminished surface localization of mutant proteins. In addition, TRPV4-regulated Ca(2+) influx was substantially reduced even after stimulation with 4alphaPDD, a TRPV4 channel-specific agonist, and with hypo-osmotic solution. In summary, we describe a new hereditary channelopathy caused by mutations in TRPV4 and present evidence that the resulting substitutions in the N-terminal ankyrin domain affect channel maturation, leading to reduced surface expression of functional TRPV4 channels.
Figures
Figure 1
Schematic model of TRPV4. (a) The TRPV4 protein is composed of a cytosolic N-terminal region and six transmembrane domains (green), including the pore region (magenta) and an intracellular C-terminal tail. The N-terminal region contains the ankyrin repeat domain (ARD), which consists of six ankyrin repeats (ANK), highlighted here in different colors (ANK1–6). The substitutions reported in this study are located in the outer helices of ANK4 (R269H) and ANK5 (R315W and R316C). An asterisk marks position 620 (valine). The V620I substitution results in an activating TRPV4 mutant observed in brachyolmia. The first helix (from the N terminus) corresponds to the inner helix and the second to the outer helix. (b) Ribbon diagram of the cytosolic N-terminal ARD-containing domain of TRPV4 (amino acids 111–358). The ankyrin repeats (ANK1–6) are formed by antiparallel inner and outer helices; the respective fingers are color coded. The PACSIN binding site (amino acids 142–143) is indicated with an asterisk. Here 2pnn.pdb was chosen as a template; Lishko et al..
Figure 2
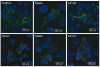
Immunolocalization of wild-type and mutant TRPV4 in transfected HeLa cells. Blue shows artificial coloring of nuclei by Hoechst; green shows anti–His tag staining. Cells were transfected with plasmid DNA (pIRES2-DsRed-Express vector) encoding His6-tagged wild-type TRPV4 (hTRPV4), three TRPV4 mutants (R269H, R315W and R316C) and two rescue mutants (R269K and R315K). Cells transfected with hTRPV4 showed localization of the protein to the plasma membrane. Mutants R269H, R315W and R316C were detected as patches distributed over the entire cytoplasm; no staining at the plasma membrane was observed. TRPV4 staining in the rescue mutants R269K and R315K was located predominantly at the plasma membrane.
Figure 3
Coexpression of wild-type and mutant TRPV4 in transfected HeLa cells. Blue shows artificial coloring of nuclei by Hoechst staining. Green shows anti–His tag staining; red shows anti–Flag tag staining. Cells were cotransfected with plasmid DNA (backbones: pIRES2-AcGFP1 vector for the wild-type and pIRES2-DsRed-Express vector for the mutants) encoding Flag-tagged hTRPV4 and the His-tagged mutants (R269H and R315W; upper panels) or the His-tagged rescue mutants (R269K and R315K; lower panels). In cells cotransfected with hTRPV4 and the mutants R269H and R315W, the Flag-tagged wild-type protein (red) was predominantly seen at the plasma membrane, whereas the His-tagged mutant protein (green) was seen in the cytoplasm. Few mixed complexes (wild-type and mutant) (yellow) were seen. The His-tagged TRPV4 protein of the rescue mutants R269K and R315K (green) was detectable at the plasma membrane.
Figure 4
Intracellular calcium changes of TRPV4-transfected HeLa cells. (a) Original traces show calcium response during hypo-osmotic challenge in HeLa cells transfected with wild-type or mutant TRPV4 channels (green, wild-type hTRPV4; light blue, R269H; red, R316C; violet, R315W). Control (dark blue) indicates a mock-transfected cell. Fl340/Fl380, ratio of fluorescence measures at 340 and 380 nm. (b) There was no difference in the basal calcium level of the transfected cells. (c) The hypo-osmotic solution–induced calcium response was significantly reduced in the mutants compared to hTRPV4 or to the rescue mutant (R269K). (d) Coexpression (+) of the mutant channels with the wild-type protein did not result in a significant difference in the Fura-2 ratio change evoked by the hypo-osmotic solution. **P < 0.01 and ***P < 0.001, indicating significant differences compared to cells expressing hTRPV4 only. hTRPV4#, double transfection with His- and Flag-tagged hTRPV4. Data are averaged values and s.d.
Figure 5
Effect of TRPV4 substitutions on TRPV4 activation by hypo-osmotic swelling or after 4α-PDD application. (a,b) Effect of hypo-osmotic challenge of the current density (pA/pF) on wild-type hTRPV4 or mutant R269H-, R315W- and R316C-transfected HeLa cells in response to a ramp protocol. (c) Pooled data from the same series as shown in a and b. Average basal inward and outward currents at −100 mV and +100 mV of HeLa cells expressing hTRPV4, mutants R269H, R315W and R316C, and rescue mutants R269K and R315K (n ≥ 4). *** indicates significant differences compared to cells expressing hTRPV4 (P < 0.001). Basal values are shown by white bars. The values of the hypo-osmotic challenge are indicated by gray bars. Mock-transfected cells were used as a control. (d) Averaged values before stimulation (white bars) and maximal values during stimulation by hypo-osmotic challenge (gray bars) of HeLa cells coexpressing mutant and wild-type channels. (e) Averaged values before stimulation (white bars) and maximal values after 4α-PDD application (gray bars) of HeLa cells expressing wild-type or mutant channels or coexpressing mutant and wild-type channels. Cotransfections are indicated in d and e with + (n ≥ 4). DsRed and GFP double-positive cells were selected for patch-clamp studies in the cotransfection studies. Numerical values are given as means ± s.e.m.
Figure 6
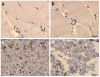
Immunohistochemistry (IHC). (a–d) Detection of TRPV4 (brown reaction product) in healthy skeletal muscle (a) and in a muscle biopsy (b) from an individual with HMSN2C (from FAM_4) harboring the TRPV4 R316C substitution. TRPV4 IHC on a nerve biopsy from a healthy control (c) and the HMSN2C patient (d). In normal tissue, TRPV4 protein is distributed in the cytoplasm (a, arrows). In the diseased tissue, reaction product is seen predominantly in the perinuclear region (b, arrows). In the healthy nerve tissue, more TRPV4-immunoreactive fibers (c, arrows) are seen than in the diseased nerve (d). Calibration bar, 10 μm.
Comment in
- Channelopathies converge on TRPV4.
Nilius B, Owsianik G. Nilius B, et al. Nat Genet. 2010 Feb;42(2):98-100. doi: 10.1038/ng0210-98. Nat Genet. 2010. PMID: 20104247 - Don't change that (calcium) channel: mutations in the same calcium channel gene can cause multiple distinct phenotypes.
Sawkins JN. Sawkins JN. Clin Genet. 2010 Aug;78(2):134-6. doi: 10.1111/j.1399-0004.2010.01452_2.x. Clin Genet. 2010. PMID: 20662855 No abstract available.
Similar articles
- Scapuloperoneal spinal muscular atrophy and CMT2C are allelic disorders caused by alterations in TRPV4.
Deng HX, Klein CJ, Yan J, Shi Y, Wu Y, Fecto F, Yau HJ, Yang Y, Zhai H, Siddique N, Hedley-Whyte ET, Delong R, Martina M, Dyck PJ, Siddique T. Deng HX, et al. Nat Genet. 2010 Feb;42(2):165-9. doi: 10.1038/ng.509. Epub 2009 Dec 27. Nat Genet. 2010. PMID: 20037587 Free PMC article. - Channelopathies converge on TRPV4.
Nilius B, Owsianik G. Nilius B, et al. Nat Genet. 2010 Feb;42(2):98-100. doi: 10.1038/ng0210-98. Nat Genet. 2010. PMID: 20104247 - Mutations in TRPV4 cause Charcot-Marie-Tooth disease type 2C.
Landouré G, Zdebik AA, Martinez TL, Burnett BG, Stanescu HC, Inada H, Shi Y, Taye AA, Kong L, Munns CH, Choo SS, Phelps CB, Paudel R, Houlden H, Ludlow CL, Caterina MJ, Gaudet R, Kleta R, Fischbeck KH, Sumner CJ. Landouré G, et al. Nat Genet. 2010 Feb;42(2):170-4. doi: 10.1038/ng.512. Epub 2009 Dec 27. Nat Genet. 2010. PMID: 20037586 Free PMC article. - TRPV4-pathy, a novel channelopathy affecting diverse systems.
Dai J, Cho TJ, Unger S, Lausch E, Nishimura G, Kim OH, Superti-Furga A, Ikegawa S. Dai J, et al. J Hum Genet. 2010 Jul;55(7):400-2. doi: 10.1038/jhg.2010.37. Epub 2010 May 27. J Hum Genet. 2010. PMID: 20505684 Review.
Cited by
- Mutations in BICD2, which encodes a golgin and important motor adaptor, cause congenital autosomal-dominant spinal muscular atrophy.
Neveling K, Martinez-Carrera LA, Hölker I, Heister A, Verrips A, Hosseini-Barkooie SM, Gilissen C, Vermeer S, Pennings M, Meijer R, te Riele M, Frijns CJ, Suchowersky O, MacLaren L, Rudnik-Schöneborn S, Sinke RJ, Zerres K, Lowry RB, Lemmink HH, Garbes L, Veltman JA, Schelhaas HJ, Scheffer H, Wirth B. Neveling K, et al. Am J Hum Genet. 2013 Jun 6;92(6):946-54. doi: 10.1016/j.ajhg.2013.04.011. Epub 2013 May 9. Am J Hum Genet. 2013. PMID: 23664116 Free PMC article. - Inherited neuropathies: clinical overview and update.
Klein CJ, Duan X, Shy ME. Klein CJ, et al. Muscle Nerve. 2013 Oct;48(4):604-22. doi: 10.1002/mus.23775. Epub 2013 Jun 26. Muscle Nerve. 2013. PMID: 23801417 Free PMC article. Review. - Improved inherited peripheral neuropathy genetic diagnosis by whole-exome sequencing.
Drew AP, Zhu D, Kidambi A, Ly C, Tey S, Brewer MH, Ahmad-Annuar A, Nicholson GA, Kennerson ML. Drew AP, et al. Mol Genet Genomic Med. 2015 Mar;3(2):143-54. doi: 10.1002/mgg3.126. Epub 2015 Jan 14. Mol Genet Genomic Med. 2015. PMID: 25802885 Free PMC article. - Defects in Axonal Transport in Inherited Neuropathies.
Beijer D, Sisto A, Van Lent J, Baets J, Timmerman V. Beijer D, et al. J Neuromuscul Dis. 2019;6(4):401-419. doi: 10.3233/JND-190427. J Neuromuscul Dis. 2019. PMID: 31561383 Free PMC article. Review. - Reduced penetrance in hereditary motor neuropathy caused by TRPV4 Arg269Cys mutation.
Berciano J, Baets J, Gallardo E, Zimoń M, García A, López-Laso E, Combarros O, Infante J, Timmerman V, Jordanova A, De Jonghe P. Berciano J, et al. J Neurol. 2011 Aug;258(8):1413-21. doi: 10.1007/s00415-011-5947-7. Epub 2011 Feb 19. J Neurol. 2011. PMID: 21336783
References
- Isozumi K, et al. Linkage of scapuloperoneal spinal muscular atrophy to chromosome 12q24.1-q24.31. Hum. Mol. Genet. 1996;5:1377–1382. - PubMed
- van der Vleuten AJ, et al. Localisation of the gene for a dominant congenital spinal muscular atrophy predominantly affecting the lower limbs to chromosome 12q23-q24. Eur. J. Hum. Genet. 1998;6:376–382. - PubMed
- McEntagart ME, et al. Confirmation of a hereditary motor and sensory neuropathy IIC locus at chromosome 12q23-q24. Ann. Neurol. 2005;57:293–297. erratum 57, 609 (2005) - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous