Inducible proteolytic inactivation of OPA1 mediated by the OMA1 protease in mammalian cells - PubMed (original) (raw)
Inducible proteolytic inactivation of OPA1 mediated by the OMA1 protease in mammalian cells
Brian Head et al. J Cell Biol. 2009.
Abstract
The mammalian mitochondrial inner membrane fusion protein OPA1 is controlled by complex patterns of alternative splicing and proteolysis. A subset of OPA1 isoforms is constitutively cleaved by YME1L. Other isoforms are not cleaved by YME1L, but they are cleaved when mitochondria lose membrane potential or adenosine triphosphate. In this study, we show that this inducible cleavage is mediated by a zinc metalloprotease called OMA1. We find that OMA1 small interfering RNA inhibits inducible cleavage, helps retain fusion competence, and slows the onset of apoptosis, showing that OMA1 controls OPA1 cleavage and function. We also find that OMA1 is normally cleaved from 60 to 40 kD by another as of yet unidentified protease. Loss of membrane potential causes 60-kD protein to accumulate, suggesting that OMA1 is attenuated by proteolytic degradation. We conclude that a proteolytic cascade controls OPA1. Inducible cleavage provides a mechanism for quality control because proteolytic inactivation of OPA1 promotes selective removal of defective mitochondrial fragments by preventing their fusion with the mitochondrial network.
Figures
Figure 1.
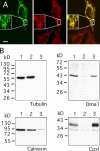
Localization of OMA1 to mitochondria in human cells. (A) HeLa cells were transfected with OMA1-HA and stained with HA antibody (left; green) and MitoTracker (middle; red). A merged image is shown on the right. Only one transfected cell is shown here, but many other transfected cells were observed, invariably showing colocalization of OMA1-HA and MitoTracker. The insets show enlargements to display the colocalization of HA antibody and MitoTracker staining more clearly. (B) Subcellular fractionation of OMA1-HA–transfected HeLa cells showing volume equivalents of postnuclear supernatant (lane 1), medium speed supernatant (lane 2), and medium speed pellet (lane 3) blotted and probed with HA, mitochondrial (COXI), and ER (calnexin) antibodies. Bar, 10 µm.
Figure 2.
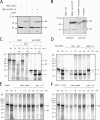
Changes in the cleavage of OMA1 upon treatment with CCCP and oligomycin. (A) HeLa cells were transfected with wild-type (wt) OMA1 or OMA1(H331A) mutant constructs. Treatment for 30 min with CCCP decreases the intensity of the 40-kD band, and a previously undetectable 60-kD protein appears. (B) Comparison between in vitro–synthesized OMA1 and the 60-kD band in OMA1-transfected and CCCP-treated HeLa cells. Unlabeled cell extract was added to radiolabeled OMA1 to load similar amounts of protein in different lanes. (A and B) Black lines indicate that intervening lanes have been spliced out. (C–F) Import of in vitro–synthesized OMA1 into isolated mitochondria. (C) Mitochondria were incubated with OMA1 or Su9-DHFR for 20 min. (D) Carbonate supernatants are shown for a Su9-DHFR time course with a 20-min carbonate (carb.) pellet as control. (E and F) Carbonate pellets are shown for an OMA1 time course with a 20-min supernatant as control. (F) Oligomycin and bongkrekic acid were added to the reactions to reduce mitochondrial ATP. IN, input lanes showing 10% of OMA1 in import reactions; m, mature protein; p, Su9-DHFR precursor; O/B, oligomycin and bongkrekic acid; Trp, trypsin; V/C, valinomycin and CCCP.
Figure 3.
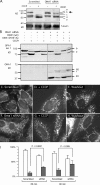
Effects of OMA1 on CCCP-induced cleavage of OPA1 and recovery of fusion competence after CCCP washout. (A) HeLa cells were transfected with OMA1 siRNA or scrambled oligonucleotides followed by 30 min with 10 µM CCCP (OMA1 RNA was reduced by 90%). CCCP-induced cleavage normally converts band b to band e, but this cleavage is inhibited by OMA1 siRNA (arrow). (B) Effects of Oma1 siRNA on CCCP-induced cleavage are suppressed by cotransfected wild-type OMA1 but not by the OMA1(H331A) mutant. Cotransfected OPA1 isoform 1 was detected with an myc tag (Griparic et al., 2007). Isoform 1 is not cleaved by YME1L, but it is cleaved by OMA1, showing a CCCP-induced shift from band b to band e. OMA1 expression and the shift from 40- to 60-kD proteins are shown on the bottom. Black lines indicate that intervening lanes have been spliced out. (C–I) Effects of OMA1 siRNA on recovery from CCCP-induced mitochondrial fragmentation. HeLa cells were transfected with scrambled (C–E) or OMA1 siRNA (F–H). Mitochondria are normally filamentous (C and F) but fragment after 30 min with CCCP (D and G). At 90 min after CCCP washout, the filamentous morphology of mitochondria returns more quickly in OMA1 siRNA cells (H) than in control cells (E). (I) Percentages of cells with fully fragmented mitochondria (open bars) or with partial or full recovery of tubular mitochondria (closed bars) are shown at 60 or 90 min after washout. Means and SD were determined with six plates in two independent experiments (400 cells per plate). P-values were determined with an unpaired Student's t test. OMA1 expression was reduced by 70% (means with real-time PCR). Bar, 10 µm.
Figure 4.
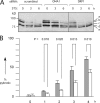
Effects of OMA1 siRNA on apoptosis. (A) HeLa cells were transfected with siRNA oligonucleotides followed by 0, 3, and 6 h with staurosporine (STS). Extracts were blotted and probed with OPA1 antibody. Staurosporine converts band b to band e, similar to CCCP-induced cleavage. OMA1 siRNA slows this process, whereas DRP1 siRNA does not, even though DRP1 siRNA inhibits apoptosis (detected with PARP and caspase cleavage; not depicted). The black line indicates that intervening lanes have been spliced out. (B) The effects of OMA1 siRNA on staurosporine-induced apoptosis were determined by counting the numbers of pyknotic nuclei in 300 cells for each time point. Mean values for three independent experiments are given with SD. The significance of differences between scrambled (closed bars) and OMA1 siRNA (open bars) was determined with an unpaired Student's t test.
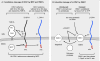
Figure 5.
Pathways for proteolytic processing of OPA1. (A) The constitutive cleavage pathway. All isoforms of OPA1 have a mitochondrial leader sequence, which is constitutively cleaved by the matrix protease mitochondrial processing peptidase (MPP). Some isoforms contain exon 4b, which has a cleavage site that we name S3, and some isoforms have exon 5b, which has a cleavage site named S2 (Ishihara et al., 2006). Isoforms with exon 4b or 5b are constitutively cleaved by YME1L to produce S-OPA1. Isoforms lacking exons 4b and 5b are not cleaved by YME1L, thus producing L-OPA1. Mitochondrial fusion requires a mixture of S- and L-OPA1. (B) The pathway for inducible cleavage. L-OPA1, which is not cleaved by YME1L, remains susceptible to inducible cleavage in the S1 site of exon 5 by OMA1. This inducible cleavage occurs when mitochondria lose membrane potential or ATP. Loss of membrane potential also leads to an accumulation of 60-kD OMA1 and reduced amounts of 40-kD OMA1, suggesting that OMA1 activity is regulated by OMA1 turnover. Another, as of yet unknown, protease may attenuate OMA1 activity by proteolytic degradation. This attenuation is slowed when mitochondria lose membrane potential or ATP, thus allowing for cleavage of OPA1. Alternatively, membrane potential or ATP levels may also directly affect OMA1 activity. IMS, intermembrane space; PD, protease domain.
Similar articles
- Membrane depolarization activates the mitochondrial protease OMA1 by stimulating self-cleavage.
Zhang K, Li H, Song Z. Zhang K, et al. EMBO Rep. 2014 May;15(5):576-85. doi: 10.1002/embr.201338240. Epub 2014 Apr 9. EMBO Rep. 2014. PMID: 24719224 Free PMC article. - Regulation of the mitochondrial dynamin-like protein Opa1 by proteolytic cleavage.
Griparic L, Kanazawa T, van der Bliek AM. Griparic L, et al. J Cell Biol. 2007 Aug 27;178(5):757-64. doi: 10.1083/jcb.200704112. Epub 2007 Aug 20. J Cell Biol. 2007. PMID: 17709430 Free PMC article. - Regulation of OPA1 processing and mitochondrial fusion by m-AAA protease isoenzymes and OMA1.
Ehses S, Raschke I, Mancuso G, Bernacchia A, Geimer S, Tondera D, Martinou JC, Westermann B, Rugarli EI, Langer T. Ehses S, et al. J Cell Biol. 2009 Dec 28;187(7):1023-36. doi: 10.1083/jcb.200906084. J Cell Biol. 2009. PMID: 20038678 Free PMC article. - Targeted OMA1 therapies for cancer.
Alavi MV. Alavi MV. Int J Cancer. 2019 Nov 1;145(9):2330-2341. doi: 10.1002/ijc.32177. Epub 2019 Feb 21. Int J Cancer. 2019. PMID: 30714136 Review. - OPA1 processing in cell death and disease - the long and short of it.
MacVicar T, Langer T. MacVicar T, et al. J Cell Sci. 2016 Jun 15;129(12):2297-306. doi: 10.1242/jcs.159186. Epub 2016 May 17. J Cell Sci. 2016. PMID: 27189080 Review.
Cited by
- Mitochondria: in sickness and in health.
Nunnari J, Suomalainen A. Nunnari J, et al. Cell. 2012 Mar 16;148(6):1145-59. doi: 10.1016/j.cell.2012.02.035. Cell. 2012. PMID: 22424226 Free PMC article. Review. - Effects of Diabetes on Mitochondrial Morphology and Its Implications in Diabetic Retinopathy.
Kim D, Roy S. Kim D, et al. Invest Ophthalmol Vis Sci. 2020 Aug 3;61(10):10. doi: 10.1167/iovs.61.10.10. Invest Ophthalmol Vis Sci. 2020. PMID: 32756920 Free PMC article. - Solving a 50 year mystery of a missing OPA1 mutation: more insights from the first family diagnosed with autosomal dominant optic atrophy.
Fuhrmann N, Schimpf S, Kamenisch Y, Leo-Kottler B, Alexander C, Auburger G, Zrenner E, Wissinger B, Alavi MV. Fuhrmann N, et al. Mol Neurodegener. 2010 Jun 14;5:25. doi: 10.1186/1750-1326-5-25. Mol Neurodegener. 2010. PMID: 20546606 Free PMC article. - Mitochondrial fission and fusion and their roles in the heart.
Kane LA, Youle RJ. Kane LA, et al. J Mol Med (Berl). 2010 Oct;88(10):971-9. doi: 10.1007/s00109-010-0674-6. Epub 2010 Sep 14. J Mol Med (Berl). 2010. PMID: 20835916 Review. - Mitochondrial form and function.
Friedman JR, Nunnari J. Friedman JR, et al. Nature. 2014 Jan 16;505(7483):335-43. doi: 10.1038/nature12985. Nature. 2014. PMID: 24429632 Free PMC article. Review.
References
- Cassidy-Stone A., Chipuk J.E., Ingerman E., Song C., Yoo C., Kuwana T., Kurth M.J., Shaw J.T., Hinshaw J.E., Green D.R., Nunnari J. 2008. Chemical inhibition of the mitochondrial division dynamin reveals its role in Bax/Bak-dependent mitochondrial outer membrane permeabilization. Dev. Cell. 14:193–204 10.1016/j.devcel.2007.11.019 - DOI - PMC - PubMed
- Cipolat S., Rudka T., Hartmann D., Costa V., Serneels L., Craessaerts K., Metzger K., Frezza C., Annaert W., D'Adamio L., et al. 2006. Mitochondrial rhomboid PARL regulates cytochrome c release during apoptosis via OPA1-dependent cristae remodeling. Cell. 126:163–175 10.1016/j.cell.2006.06.021 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases