Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination - PubMed (original) (raw)
Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination
Simon Myers et al. Science. 2010.
Abstract
Although present in both humans and chimpanzees, recombination hotspots, at which meiotic crossover events cluster, differ markedly in their genomic location between the species. We report that a 13-base pair sequence motif previously associated with the activity of 40% of human hotspots does not function in chimpanzees and is being removed by self-destructive drive in the human lineage. Multiple lines of evidence suggest that the rapidly evolving zinc-finger protein PRDM9 binds to this motif and that sequence changes in the protein may be responsible for hotspot differences between species. The involvement of PRDM9, which causes histone H3 lysine 4 trimethylation, implies that there is a common mechanism for recombination hotspots in eukaryotes but raises questions about what forces have driven such rapid change.
Figures
Figure 1
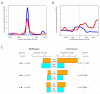
Recombination rates and patterns of motif gain and loss in human and chimpanzee. For additional details, see (8). A Estimated HapMap Phase II recombination rate across the 40kb surrounding 16 human THE1 elements (red line) and 6 L2 elements (blue line) orthologous to the 22 regions analyzed in chimpanzee, and each containing a conserved exact match to the 13-bp core motif. Rates are smoothed using a 2kb sliding window slid in 50bp increments, averaged across elements. Horizontal dashed line: the human average recombination rate of 1.1cM/Mb. Vertical dotted line: the centre of the repeat. B Average estimated recombination rate for the western chimpanzee data across around the 16 THE1 elements (red line) and six L2 elements (blue line) containing the 13-bp core motif. Other details as for A. C Numbers of core motif gains (left hand bars) versus losses (right bars), inferred using macaque and orang-utan outgroup information (8), in humans (orange bars) and chimpanzees (cyan bars) on three backgrounds; THE1, L2 and non-repeat (NR). For each background, gains are shown as a fraction of motifs currently present in each species, losses as a fraction of motifs inferred in the human-chimpanzee ancestor. The intervals flanking the plot on each side show exact 1-sided 95% confidence intervals and associated p-values for testing equality of gain/loss rate between the species (8).
Figure 2
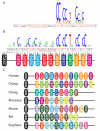
A Previously estimated degeneracy of the 13-bp hotspot motif (6) (logo plot; relative letter height proportional to estimated probability of hotspot activity, total letter height determined by degree of base specificity) as well as an extended ~39-bp motif (text below logo, with influential positions (p<0.01) shown in red). B In silico prediction of the binding consensus for PRDM9, aligned with the 13-mer, with more influential positions shown in red. Underlined in both A and B is an additional 8-bp matching sequence. The logo shows predicted degeneracy within this consensus (8). Below the text is the sequence of four predicted DNA-contacting amino acids for the 13 successive human PRDM9 zinc fingers (1 oval per finger, differing colors for differing fingers, separated finger is gapped N-terminal from others), and their predicted base contacts within the motif. C Sequence of four predicted DNA-contacting amino acids for the PRDM9 zinc fingers in 7 mammalian species, presented as in B. Distinct fingers given different colors; fingers present in at least two species have black border.
Comment in
- Genetics. Genetic control of hotspots.
Cheung VG, Sherman SL, Feingold E. Cheung VG, et al. Science. 2010 Feb 12;327(5967):791-2. doi: 10.1126/science.1187155. Science. 2010. PMID: 20150474 No abstract available.
Similar articles
- The red queen model of recombination hotspots evolution in the light of archaic and modern human genomes.
Lesecque Y, Glémin S, Lartillot N, Mouchiroud D, Duret L. Lesecque Y, et al. PLoS Genet. 2014 Nov 13;10(11):e1004790. doi: 10.1371/journal.pgen.1004790. eCollection 2014 Nov. PLoS Genet. 2014. PMID: 25393762 Free PMC article. - Mouse PRDM9 DNA-binding specificity determines sites of histone H3 lysine 4 trimethylation for initiation of meiotic recombination.
Grey C, Barthès P, Chauveau-Le Friec G, Langa F, Baudat F, de Massy B. Grey C, et al. PLoS Biol. 2011 Oct;9(10):e1001176. doi: 10.1371/journal.pbio.1001176. Epub 2011 Oct 18. PLoS Biol. 2011. PMID: 22028627 Free PMC article. - PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice.
Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C, Przeworski M, Coop G, de Massy B. Baudat F, et al. Science. 2010 Feb 12;327(5967):836-40. doi: 10.1126/science.1183439. Epub 2009 Dec 31. Science. 2010. PMID: 20044539 Free PMC article. - PRDM9 and Its Role in Genetic Recombination.
Paigen K, Petkov PM. Paigen K, et al. Trends Genet. 2018 Apr;34(4):291-300. doi: 10.1016/j.tig.2017.12.017. Epub 2018 Jan 21. Trends Genet. 2018. PMID: 29366606 Free PMC article. Review. - The case of the fickle fingers: how the PRDM9 zinc finger protein specifies meiotic recombination hotspots in humans.
Ségurel L, Leffler EM, Przeworski M. Ségurel L, et al. PLoS Biol. 2011 Dec;9(12):e1001211. doi: 10.1371/journal.pbio.1001211. Epub 2011 Dec 6. PLoS Biol. 2011. PMID: 22162947 Free PMC article. Review.
Cited by
- sgcocaller and comapr: personalised haplotype assembly and comparative crossover map analysis using single-gamete sequencing data.
Lyu R, Tsui V, Crismani W, Liu R, Shim H, McCarthy DJ. Lyu R, et al. Nucleic Acids Res. 2022 Nov 11;50(20):e118. doi: 10.1093/nar/gkac764. Nucleic Acids Res. 2022. PMID: 36107768 Free PMC article. - Commercial chicken breeds exhibit highly divergent patterns of linkage disequilibrium.
Pengelly RJ, Gheyas AA, Kuo R, Mossotto E, Seaby EG, Burt DW, Ennis S, Collins A. Pengelly RJ, et al. Heredity (Edinb). 2016 Nov;117(5):375-382. doi: 10.1038/hdy.2016.47. Epub 2016 Jul 6. Heredity (Edinb). 2016. PMID: 27381324 Free PMC article. - Nonhistone Lysine Methylation in the Regulation of Cancer Pathways.
Carlson SM, Gozani O. Carlson SM, et al. Cold Spring Harb Perspect Med. 2016 Nov 1;6(11):a026435. doi: 10.1101/cshperspect.a026435. Cold Spring Harb Perspect Med. 2016. PMID: 27580749 Free PMC article. Review. - The molecular machinery of meiotic recombination.
Chen L, Weir JR. Chen L, et al. Biochem Soc Trans. 2024 Feb 28;52(1):379-393. doi: 10.1042/BST20230712. Biochem Soc Trans. 2024. PMID: 38348856 Free PMC article. Review. - Genomic variation in natural populations of Drosophila melanogaster.
Langley CH, Stevens K, Cardeno C, Lee YC, Schrider DR, Pool JE, Langley SA, Suarez C, Corbett-Detig RB, Kolaczkowski B, Fang S, Nista PM, Holloway AK, Kern AD, Dewey CN, Song YS, Hahn MW, Begun DJ. Langley CH, et al. Genetics. 2012 Oct;192(2):533-98. doi: 10.1534/genetics.112.142018. Epub 2012 Jun 5. Genetics. 2012. PMID: 22673804 Free PMC article.
References
- McVean GA, et al. Science. 2004;304:581. - PubMed
- Ptak SE, et al. Nat Genet. 2005;37:429. - PubMed
- Winckler W, et al. Science. 2005;308:107. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases