Epigenetic regulation of latent HSV-1 gene expression - PubMed (original) (raw)
Review
Epigenetic regulation of latent HSV-1 gene expression
David C Bloom et al. Biochim Biophys Acta. 2010 Mar-Apr.
Abstract
Like other alpha-herpesviruses, Herpes Simplex Virus Type 1 (HSV-1) possesses the ability to establish latency in sensory ganglia as a non-integrated, nucleosome-associated episome in the host cell nucleus. Transcription of the genome is limited to the Latency-Associated Transcript (LAT), while the lytic genes are maintained in a transcriptionally repressed state. This partitioning of the genome into areas of active and inactive transcription suggests epigenetic control of HSV-1 latent gene expression. During latency viral transcription is not regulated by DNA methylation but likely by post-translational histone modifications. The LAT region is the only region of the genome enriched in marks indicative of transcriptional permissiveness, specifically dimethyl H3 K4 and acetyl H3 K9, K14, while the lytic genes appear under-enriched in those same marks. In addition, facultative heterochromatin marks, specifically trimethyl H3 K27 and the histone variant macroH2A, are enriched on lytic genes during latency. The distinct epigenetic domains of the LAT and the lytic genes appear to be separated by chromatin insulators. Binding of CTCF, a protein that binds to all known vertebrate insulators, to sites within the HSV-1 genome likely prevents heterochromatic spreading and blocks enhancer activity. When the latent viral genome undergoes stress-induced reactivation, it is possible that CTCF binding and insulator function are abrogated, enabling lytic gene transcription to ensue. In this review we summarize our current understanding of latent HSV-1 epigenetic regulation as it pertains to infections in both the rabbit and mouse models. CTCF insulator function and regulation of histone tail modifications will be discussed. We will also present a current model of how the latent genome is carefully controlled at the epigenetic level and how stress-induced changes to it may trigger reactivation.
2009 Elsevier B.V. All rights reserved.
Figures
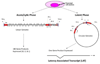
Figure 1. Stages of an HSV-1 infection in vivo
Humans typically acquire HSV-1 as a primary infection of the mucosa of the lips or eye. During the initial acute infection, the virus replicates locally in the mucosal epithelium and gains access to the sensory nerve termini that underlie the skin’s surface. The virus then travels to the nerve cell bodies in the trigeminal ganglia where the virus will replicate in some cells and become latent in others. During the latent infection, the virus remains dormant in the trigeminal ganglion. During this period no infectious virus is detected, and viral gene expression is suppressed, except for the latency-associated transcript (LAT). Periodically, stress causes the virus to reactivate. During reactivation viral lytic gene transcription and DNA replication initiates in some neurons, and virions are transported back down the axons to the primary site of infection. This results in infectious virus at the site of initial infection, and in some instances, clinical lesions such as cold sores will result.
Figure 2. Molecular features of HSV-1 lytic and latent infections
Once HSV-1 enters a sensory neuron, it can either initiate lytic replication or establish a latent infection. The acute or lytic phase is characterized by expression of all three kinetic gene classes (immediate early, IE; early, E; or late, L) and replication of the viral genome through a linear mechanism. In contrast, the latent infection is characterized by circularization of the viral genome and suppression of the lytic genes. Only one gene product, the non-coding LAT, is abundantly transcribed during latency. This figure also illustrates the four different regions of the HSV-1 genome: the unique long (UL) region, the unique short (US) region, and the two sets of repeats, the repeat long (RL) and the repeat short (RS). Each of the repeats is present in two copies, differentiated as either the terminal (T) or internal (I) segment.
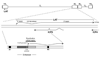
Figure 3. Features of the LAT region
The LAT exists in duplicate in the genome, since it is located in the long repeat (RL) regions. The 8.5 kb LAT primary transcript is shown, with the exons and 2.0 kb stable intron indicated. Additionally, the lytic ICP0 and ICP4 transcripts are designated. The lower blown-up section gives additional key features of the LAT that are relevant to this review, specifically those in the region upstream of the LAT through the LAT intron. The B1 and B2 insulators are indicated by black boxes; the LAT promoter, core promoter, enhancer, and reactivation critical region are also shown. The LAT transcriptional start site is designated by a filled black arrow.
Figure 4. Chromatin profile of the latent HSV-1 genome
A) Regions in which euchromatic histone marks are enriched are denoted by green stars on the HSV-1 genome. B) Regions in which heterochromatic histone marks are enriched are marked in purple boxes on the HSV-1 genome. C) The enlarged LAT region displays the areas where euchromatic (acetyl H3 K9, K14 and dimethyl H3 K4) and heterochromatic (trimethyl H3 K27 and trimethyl H3 K9) marks were identified.
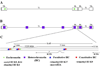
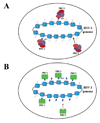
Figure 5. Potential chromatin populations of latent HSV-1 genomes
A) Genomes that associate with trimethyl H3 K27 (blue triangles) are present in different cells than genomes associated with trimethyl H3 K9 (red circles). B) Genomes enriched in either trimethyl H3K27 or trimethyl H3 K9 are present within the same cell in latent ganglia. C) Genes in the viral genome present in duplicate have differing histone post-translational marks on the two different copies of the same gene.
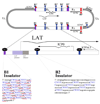
Figure 6. Location of CTCF binding sites in the HSV-1 genome
The HSV-1 genome contains 7 unique clusters of CTCF binding sites. CTCF-binding sites are often present in clusters, and the cluster CTRL2 (of the B2 insulator) located 3’ to the LAT enhancer contains 9 copies of the consensus, whereas the cluster CTRL1 upstream of the LAT promoter (of the B1 insulator) contains 46 copies, in total, of two separate consensus motifs, CTCCC (blue), and CCCTC (underlined in red). These CTCF binding sites have been shown to bind the cellular insulator protein CTCF during latency. The B1 and B2 insulators flank the only region of the genome that is marked with transcriptionally permissive histone modifications during latency. It is speculated that these two insulators act as boundary elements to segregate the transcriptionally active LAT promoter regions from the repressed regions of the genome, as well as to prevent the LAT enhancer from acting on the surrounding lytic genes.
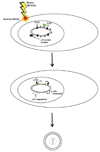
Figure 7. Model of polycomb-mediated repression on the latent HSV-1 genome
A) PRC2 protein complexes are recruited to the genome and trimethylate H3K27. B) PRC1 replaces PRC2 on the genome to maintain repressive histone marks to keep the latent genome in a transcriptionally inhibited state.
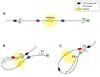
Figure 8. CTCF insulators can interact to regulate transcription through the formation of loop domains that can direct the formation of transcriptional hubs
A) Chromatin insulators, with CTCF bound, can act as enhancer-blockers to prevent the enhancer from activating genes located distal to the insulators. B) Interactions between the CTCF proteins bound to the blue and black insulators form a loop domain that allows the P1 promoter to be influenced by the enhancer. C) Interaction between the CTCFs on the green and red insulator elements excludes the P1 promoter from activation, but now allows the P2 promoter to be activated by the enhancer.
Figure 9. Model for reversal of epigenetic repression of lytic genes during HSV reactivation
During latency, the LAT acts to balance transcriptionally repressive (but reversible) facultative heterochromatin marks (purple boxes) on the lytic gene regions of the genome. These heterochromatic marks are established and maintained by polycomb group proteins (green rectangle). The CTCF insulators (black boxes) flanking that LAT promoter/enhancer block the LAT enhancer from activating ICP0 and other lytic genes. Stressors and other reactivation mediators cause a transient decrease in the LAT, and possibly a loss of insulator occupancy at the B2 and/or B1 insulators. This allows the LAT enhancer to activate lytic genes, resulting in viral replication and the production of infectious virus.
Similar articles
- The polycomb group protein Bmi1 binds to the herpes simplex virus 1 latent genome and maintains repressive histone marks during latency.
Kwiatkowski DL, Thompson HW, Bloom DC. Kwiatkowski DL, et al. J Virol. 2009 Aug;83(16):8173-81. doi: 10.1128/JVI.00686-09. Epub 2009 Jun 10. J Virol. 2009. PMID: 19515780 Free PMC article. - The CCCTC Binding Factor, CTRL2, Modulates Heterochromatin Deposition and the Establishment of Herpes Simplex Virus 1 Latency In Vivo.
Washington SD, Singh P, Johns RN, Edwards TG, Mariani M, Frietze S, Bloom DC, Neumann DM. Washington SD, et al. J Virol. 2019 Jun 14;93(13):e00415-19. doi: 10.1128/JVI.00415-19. Print 2019 Jul 1. J Virol. 2019. PMID: 30996085 Free PMC article. - Chromatin-mediated epigenetic regulation of HSV-1 transcription as a potential target in antiviral therapy.
Schang LM, Hu M, Cortes EF, Sun K. Schang LM, et al. Antiviral Res. 2021 Aug;192:105103. doi: 10.1016/j.antiviral.2021.105103. Epub 2021 Jun 1. Antiviral Res. 2021. PMID: 34082058 Free PMC article. Review. - Epigenotypes of latent herpesvirus genomes.
Minarovits J. Minarovits J. Curr Top Microbiol Immunol. 2006;310:61-80. doi: 10.1007/3-540-31181-5_5. Curr Top Microbiol Immunol. 2006. PMID: 16909907 Review.
Cited by
- Herpes simplex virus 1 DNA is in unstable nucleosomes throughout the lytic infection cycle, and the instability of the nucleosomes is independent of DNA replication.
Lacasse JJ, Schang LM. Lacasse JJ, et al. J Virol. 2012 Oct;86(20):11287-300. doi: 10.1128/JVI.01468-12. Epub 2012 Aug 8. J Virol. 2012. PMID: 22875975 Free PMC article. - Epigenetic repression of herpes simplex virus infection by the nucleosome remodeler CHD3.
Arbuckle JH, Kristie TM. Arbuckle JH, et al. mBio. 2014 Jan 14;5(1):e01027-13. doi: 10.1128/mBio.01027-13. mBio. 2014. PMID: 24425734 Free PMC article. - Inhibition of the Super Elongation Complex Suppresses Herpes Simplex Virus Immediate Early Gene Expression, Lytic Infection, and Reactivation from Latency.
Alfonso-Dunn R, Arbuckle JH, Vogel JL, Kristie TM. Alfonso-Dunn R, et al. mBio. 2020 Jun 9;11(3):e01216-20. doi: 10.1128/mBio.01216-20. mBio. 2020. PMID: 32518191 Free PMC article. - Deletion of Herpes Simplex Virus 1 MicroRNAs miR-H1 and miR-H6 Impairs Reactivation.
Barrozo ER, Nakayama S, Singh P, Vanni EAH, Arvin AM, Neumann DM, Bloom DC. Barrozo ER, et al. J Virol. 2020 Jul 16;94(15):e00639-20. doi: 10.1128/JVI.00639-20. Print 2020 Jul 16. J Virol. 2020. PMID: 32295910 Free PMC article. - Occult hepatitis B virus and hepatocellular carcinoma.
Pollicino T, Saitta C. Pollicino T, et al. World J Gastroenterol. 2014 May 28;20(20):5951-61. doi: 10.3748/wjg.v20.i20.5951. World J Gastroenterol. 2014. PMID: 24876718 Free PMC article. Review.
References
- Cook SD, Ophth FC, Hill JH. Herpes simplex virus: molecular biology and the possibility of corneal latency. Survey of Ophthalmol. 1991;36(2):140–148. - PubMed
- Clements GB, Jamieson FE. Reactivation of latent herpes simplex virus-1 (HSV) from mouse footpad cells demonstrated by in situ hybridization. Arch Virol. 1989;104(1–2):95–106. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI048633/AI/NIAID NIH HHS/United States
- R01 AI048633-08/AI/NIAID NIH HHS/United States
- T32 AI007110/AI/NIAID NIH HHS/United States
- T32 AI007110-26A2/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials