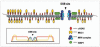The complexity of phosphorylated H2AX foci formation and DNA repair assembly at DNA double-strand breaks - PubMed (original) (raw)
The complexity of phosphorylated H2AX foci formation and DNA repair assembly at DNA double-strand breaks
Asako J Nakamura et al. Cell Cycle. 2010.
Abstract
The maintenance of genome stability requires efficient DNA double-stranded break (DSB) repair mediated by the phosphorylation of multiple histone H2AX molecules near the break sites. The phosphorylated H2AX (gammaH2AX) molecules form foci covering many megabases of chromatin. The formation of gamma-H2AX foci is critical for efficient DNA damage response (DDR) and for the maintenance of genome stability, however, the mechanisms of protein organization in foci is largely unknown. To investigate the nature of gammaH2AX foci formation, we analyzed the distribution of gammaH2AX and other DDR proteins at DSB sites using a variety of techniques to visualize, expand and partially disrupt chromatin. We report here that gammaH2AX foci change composition during the cell cycle, with proteins 53BP1, NBS1 and MRE11 dissociating from foci in G(2) and mitosis to return at the beginning of the following G(1). In contrast, MDC1 remained colocalized with gamma-H2AX during mitosis. In addition, while gammaH2AX was found to span large domains flanking DSB sites, 53BP1 and NBS1 were more localized and MDC1 colocalized in doublets in foci. H2AX and MDC1 were found to be involved in chromatin relaxation after DSB formation. Our data demonstrates that the DSB repair focus is a heterogeneous and dynamic structure containing internal complexity.
Figures
Figure 1
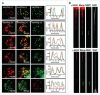
The components of IRIF are reorganized during the cell cycle. HeLa cells were synchronized by colcemid, exposed to 1 Gy IR and fixed after 30 min incubation. Mitotic stages were classified by DAPI staining. (A) Immunostaining for γH2AX and 53BP1. I-III refer to enlargements of the noted areas. (B) Immunostaining for γH2AX and MDC1. (C) Fraction of γH2AX foci colocalized with 53BP1 (left) or MDC1 (right).
Figure 2
γH2AX foci enlarge upon chromatin swelling as do MDC1 foci, while 53BP1, NBS1 and MRE11 foci do not. (A, left images) At 30 min after exposure to 1 Gy, HeLa cells were subjected to swelling, then double-immunostained for γH2AX and 53BP1, MRE11, NBS1 or MDC1. The top row shows a no-swelling control with 53BP1. The bottom row shows a reversed-staining control. (A, right graphs) Density scanning along the white line shown in the merged images. (B) Immunostaining of γH2AX and either 53BP1 or NBS1 on chromatin fibers from Indian Muntjac cells. Chromatin fibers were prepared at 30 min after 1 Gy-exposure and immunostained for γH2AX and either 53BP1 or NBS1.
Figure 3
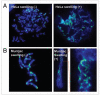
γH2AX foci enlarge on swollen metaphase chromosomes. (A) HeLa metaphase chromosomes immunostained for γH2AX with (right) or without (left) swelling. The metaphase spreads from HeLa cells were incubated with sterile water at room temperature for 15 min. After the solution was carefully removed from the cells, they were fixed by 80% ethanol at −20°C for 30 min. (B) Indian Muntjac cell metaphase chromosomes immunostained for γH2AX after (right) or without (left) swelling. The right panel shows a greater magnification of the noted area in the middle panel.
Figure 4
Until 2 hr post IR, γH2AX localizes to a large region around the DNA DSB site. (A, left images) Swollen interphase nuclei were prepared from HeLa cells at the noted times after 1 Gy-exposure and immunostained for γH2AX and 53BP1. (A, right graphs) Density scanning along the white line shown in the merged images. Note that at 2 hr and beyond, γH2AX foci do not expand on swollen chromatin. (B) Graphical presentation of the data in (A). Average from three independent experiments. At least 25 foci were counted in each experiment. Error bars signify standard deviation.
Figure 5
MDC1 form double foci appearing to bracket γH2AX foci. (A) HeLa metaphase chromosomes stained for γH2AX and MDC1. The right panels show magnified images of the boxed areas in the merged image. (B) HeLa metaphase chromosomes stained for γH2AX and MDC1 and analyzed by the DeltaVision OMX™ 3D-SIM™ Super Resolution Imaging System.
Figure 6
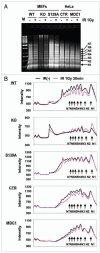
γH2AX and MDC1 are involved in chromatin relaxation after IR exposure. (A) Increased MNase accessibility of chromatin after IR exposure. Cells from H2AX wild type MEFs (WT), H2AX-null MEFs (KO), H2AX-S139A mutated H2AX MEFs (S139A), HeLa cells (CTR) and MDC1-depleted HeLa cells (MDC1 siRNA) were treated by MNase for 5 min, and the fragments analyzed by gel electrophoresis. (B) Profiles of the various lanes of the gel presented in (A). N1-N7 denote the numbers of nucleosomes per oligonucleosome. Note the shift in oligonucleosome size in wild type H2AX or MDC1 cells after IR exposure.
Figure 7
Distribution model of DNA repair proteins in foci surrounding a DSB. γH2AX form IRIF in a large area around DNA DSB site. MDC1 accumulate primarily at the outside edges of γ-H2AX focus. In contrast, 53BP1, NBS1 and MRE11 accumulate primarily in a smaller region near the DNA DSB site.
Comment in
- DNA double strand breaks: not all foci are created equal.
Cobb JA, Lees-Miller SP. Cobb JA, et al. Cell Cycle. 2010 Feb 1;9(3):442-3. Epub 2010 Feb 1. Cell Cycle. 2010. PMID: 20130448 No abstract available. - Shifting focus.
Nelson G, von Zglinicki T. Nelson G, et al. Cell Cycle. 2010 Feb 1;9(3):443-4. Epub 2010 Feb 1. Cell Cycle. 2010. PMID: 20130449 No abstract available. - The anatomy and cell cycle evolution of DNA damage signaling and repair foci.
Iliakis G. Iliakis G. Cell Cycle. 2010 Feb 1;9(3):444-5. Epub 2010 Feb 1. Cell Cycle. 2010. PMID: 20130450 No abstract available.
Similar articles
- Clustered DNA damage induces pan-nuclear H2AX phosphorylation mediated by ATM and DNA-PK.
Meyer B, Voss KO, Tobias F, Jakob B, Durante M, Taucher-Scholz G. Meyer B, et al. Nucleic Acids Res. 2013 Jul;41(12):6109-18. doi: 10.1093/nar/gkt304. Epub 2013 Apr 24. Nucleic Acids Res. 2013. PMID: 23620287 Free PMC article. - Wild-type p53-induced phosphatase 1 dephosphorylates histone variant gamma-H2AX and suppresses DNA double strand break repair.
Moon SH, Lin L, Zhang X, Nguyen TA, Darlington Y, Waldman AS, Lu X, Donehower LA. Moon SH, et al. J Biol Chem. 2010 Apr 23;285(17):12935-47. doi: 10.1074/jbc.M109.071696. Epub 2010 Jan 29. J Biol Chem. 2010. PMID: 20118229 Free PMC article. - 53BP1-dependent robust localized KAP-1 phosphorylation is essential for heterochromatic DNA double-strand break repair.
Noon AT, Shibata A, Rief N, Löbrich M, Stewart GS, Jeggo PA, Goodarzi AA. Noon AT, et al. Nat Cell Biol. 2010 Feb;12(2):177-84. doi: 10.1038/ncb2017. Epub 2010 Jan 17. Nat Cell Biol. 2010. PMID: 20081839 - The influence of heterochromatin on DNA double strand break repair: Getting the strong, silent type to relax.
Goodarzi AA, Jeggo P, Lobrich M. Goodarzi AA, et al. DNA Repair (Amst). 2010 Dec 10;9(12):1273-82. doi: 10.1016/j.dnarep.2010.09.013. Epub 2010 Oct 30. DNA Repair (Amst). 2010. PMID: 21036673 Review. - Double strand break repair functions of histone H2AX.
Scully R, Xie A. Scully R, et al. Mutat Res. 2013 Oct;750(1-2):5-14. doi: 10.1016/j.mrfmmm.2013.07.007. Epub 2013 Jul 31. Mutat Res. 2013. PMID: 23916969 Free PMC article. Review.
Cited by
- Vimentin protects cells against nuclear rupture and DNA damage during migration.
Patteson AE, Vahabikashi A, Pogoda K, Adam SA, Mandal K, Kittisopikul M, Sivagurunathan S, Goldman A, Goldman RD, Janmey PA. Patteson AE, et al. J Cell Biol. 2019 Dec 2;218(12):4079-4092. doi: 10.1083/jcb.201902046. Epub 2019 Nov 1. J Cell Biol. 2019. PMID: 31676718 Free PMC article. - 89Zr-PET imaging of DNA double-strand breaks for the early monitoring of response following α- and β-particle radioimmunotherapy in a mouse model of pancreatic ductal adenocarcinoma.
Poty S, Mandleywala K, O'Neill E, Knight JC, Cornelissen B, Lewis JS. Poty S, et al. Theranostics. 2020 Apr 27;10(13):5802-5814. doi: 10.7150/thno.44772. eCollection 2020. Theranostics. 2020. PMID: 32483420 Free PMC article. - Discovery of Potent and Selective MTH1 Inhibitors for Oncology: Enabling Rapid Target (In)Validation.
Farand J, Kropf JE, Blomgren P, Xu J, Schmitt AC, Newby ZE, Wang T, Murakami E, Barauskas O, Sudhamsu J, Feng JY, Niedziela-Majka A, Schultz BE, Schwartz K, Viatchenko-Karpinski S, Kornyeyev D, Kashishian A, Fan P, Chen X, Lansdon EB, Ports MO, Currie KS, Watkins WJ, Notte GT. Farand J, et al. ACS Med Chem Lett. 2019 Nov 19;11(3):358-364. doi: 10.1021/acsmedchemlett.9b00420. eCollection 2020 Mar 12. ACS Med Chem Lett. 2019. PMID: 32184970 Free PMC article. - Activation of DNA damage response signaling by condensed chromatin.
Burgess RC, Burman B, Kruhlak MJ, Misteli T. Burgess RC, et al. Cell Rep. 2014 Dec 11;9(5):1703-1717. doi: 10.1016/j.celrep.2014.10.060. Epub 2014 Nov 20. Cell Rep. 2014. PMID: 25464843 Free PMC article. - Recruitment of cyclin G2 to promyelocytic leukemia nuclear bodies promotes dephosphorylation of γH2AX following treatment with ionizing radiation.
Naito Y, Yabuta N, Sato J, Ohno S, Sakata M, Kasama T, Ikawa M, Nojima H. Naito Y, et al. Cell Cycle. 2013 Jun 1;12(11):1773-84. doi: 10.4161/cc.24878. Epub 2013 May 8. Cell Cycle. 2013. PMID: 23656780 Free PMC article.
References
- Fernandez-Capetillo O, Celeste A, Nussenzweig A. Focusing on foci: H2AX and the recruitment of DNA-damage response factors. Cell Cycle. 2003;2:426–7. - PubMed
- Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol. 2000;10:886–95. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous