NMR mapping of the IFNAR1-EC binding site on IFNalpha2 reveals allosteric changes in the IFNAR2-EC binding site - PubMed (original) (raw)
NMR mapping of the IFNAR1-EC binding site on IFNalpha2 reveals allosteric changes in the IFNAR2-EC binding site
Sabine Ruth Akabayov et al. Biochemistry. 2010.
Abstract
All type I interferons (IFNs) bind to a common cell-surface receptor consisting of two subunits. IFNs initiate intracellular signal transduction cascades by simultaneous interaction with the extracellular domains of its receptor subunits, IFNAR1 and IFNAR2. In this study, we mapped the surface of IFNalpha2 interacting with the extracellular domain of IFNAR1 (IFNAR1-EC) by following changes in or the disappearance of the (1)H-(15)N TROSY-HSQC cross peaks of IFNalpha2 caused by the binding of the extracellular domain of IFNAR1 (IFNAR1-EC) to the binary complex of IFNalpha2 with IFNAR2-EC. The NMR study of the 89 kDa complex was conducted at pH 8 and 308 K using an 800 MHz spectrometer. IFNAR1 binding affected a total of 47 of 165 IFNalpha2 residues contained in two large patches on the face of the protein opposing the binding site for IFNAR2 and in a third patch located on the face containing the IFNAR2 binding site. The first two patches form the IFNAR1 binding site, and one of these matches the IFNAR1 binding site previously identified by site-directed mutagenesis. The third patch partially matches the IFNalpha2 binding site for IFNAR2-EC, indicating allosteric communication between the binding sites for the two receptor subunits.
Figures
FIGURE 1
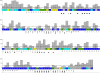
Overlay of the [15N,1H]-TROSY-HSQC spectra of IFNα2/IFNAR2-EC binary complex (black) and IFNAR1-EC/IFNα2/IFNAR2-EC ternary complex (red) (A) and expansion of the central (boxed) region of this spectrum (B). Residues that did not change their chemical shift significantly upon IFNAR1-EC binding are labeled in blue, residues that underwent significant changes in chemical shift upon IFNAR1-EC binding are labeled in yellow (0.05<Δδ<0.10 ppm); residues that could not be assigned due to large changes in chemical shift or disappearance upon IFNAR1-EC binding are labeled in black. Spectra were measured at 308 K and pH 8.
FIGURE 2
A summary of the changes in IFNα2 HSQC cross peaks upon IFNAR1-EC binding to the binary IFNα2/IFNAR2-EC complex. Residues located in helices are marked with the corresponding helix label (A, B, C, D and E). Residues that did not change their chemical shift significantly upon IFNAR1-EC binding are colored in deep blue, residues that underwent significant chemical shift changes upon IFNAR1-EC binding are colored in yellow (0.05<Δδ<0.10 ppm); residues that could not be assigned due to large chemical shift changes or disappearance upon IFNAR1-EC binding are colored in grey, overlapping cross peaks which include one cross peak that disappeared or underwent a large chemical shift change but could not be assigned due to overlap are colored green and residues whose HSQC cross peaks could not be assigned in the IFNα2/IFNAR2-EC binary complex are colored in cyan. The exposure of IFNα2 residues (backbone and sidechain included) in the binary complex are given by grey bars with a scale from 0–100%. A dotted line represents 20% exposure and below this value of exposure residues are defined as buried. Filled black triangles indicate residues that are not buried and are implicated in IFNAR1-EC binding by the present study. Buried residues that underwent significant changes in chemical shift or disappeared upon IFNAR1-EC binding and were not found to be involved in IFNAR2-EC binding are marked with asterisks. Filled black circles indicate residues found by NMR and site directed mutagenesis to interact with IFNAR2-EC in the binary complex (4, 22, 24).
FIGURE 3
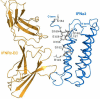
Mapping of the residues that exhibited significant chemical shift changes or disappeared upon IFNAR1-EC binding on the surface of IFNα2. A) The face of IFNα2 opposing the binding site for IFNAR2-EC and that contain the two patches involved in IFNAR1-EC binding according to the present study. B) The face of IFNα2 containing the surface previously found to be involved in IFNAR2-EC binding. IFNAR2-EC bound to IFNα2 is presented by an orange ribbon diagram according to the previously calculated model of the binary complex (24). Residues previously identified by site directed mutagenesis as being involved in IFNAR1-EC binding (25) are marked with dotted purple surfaces. Other color coding is the same as in Figure 2. All molecular pictures were created using Pymol (40).
FIGURE 4
Comparison of IFNα2 (top) and IFNβ (bottom) residues implicated in IFNAR1-EC binding. Bold characters indicate IFNα2 and IFNβ residues found in the helices. Residues in boxes are IFNα2 residues implicated in IFNAR1-EC binding in the present study. Black triangles designate residues of IFNα2 that were determined by mutagenesis to be in the binding site for IFNAR1-EC (25, 29). Filled black circles indicate residues of IFNβ that were determined by mutagenesis to be in the binding site of IFNAR1-EC (28).
FIGURE 5
A side view of the IFNα2/IFNAR2-EC binding interface. The IFNα2 and the IFNAR2-EC are colored blue and orange respectively. The E-Helix and the C-terminal tail residues that disappeared upon IFNAR1-EC binding to the binary complex are shown in stick representation and in gray.
Similar articles
- Intermolecular interactions in a 44 kDa interferon-receptor complex detected by asymmetric reverse-protonation and two-dimensional NOESY.
Nudelman I, Akabayov SR, Schnur E, Biron Z, Levy R, Xu Y, Yang D, Anglister J. Nudelman I, et al. Biochemistry. 2010 Jun 29;49(25):5117-33. doi: 10.1021/bi100041f. Biochemistry. 2010. PMID: 20496919 Free PMC article. - Ligand-induced assembling of the type I interferon receptor on supported lipid bilayers.
Lamken P, Lata S, Gavutis M, Piehler J. Lamken P, et al. J Mol Biol. 2004 Jul 30;341(1):303-18. doi: 10.1016/j.jmb.2004.05.059. J Mol Biol. 2004. PMID: 15312780 - Mutational analysis of the IFNAR1 binding site on IFNalpha2 reveals the architecture of a weak ligand-receptor binding-site.
Roisman LC, Jaitin DA, Baker DP, Schreiber G. Roisman LC, et al. J Mol Biol. 2005 Oct 21;353(2):271-81. doi: 10.1016/j.jmb.2005.08.042. J Mol Biol. 2005. PMID: 16171819 - The receptor of the type I interferon family.
Uzé G, Schreiber G, Piehler J, Pellegrini S. Uzé G, et al. Curr Top Microbiol Immunol. 2007;316:71-95. doi: 10.1007/978-3-540-71329-6_5. Curr Top Microbiol Immunol. 2007. PMID: 17969444 Review. - Interferon Receptor Trafficking and Signaling: Journey to the Cross Roads.
Zanin N, Viaris de Lesegno C, Lamaze C, Blouin CM. Zanin N, et al. Front Immunol. 2021 Jan 20;11:615603. doi: 10.3389/fimmu.2020.615603. eCollection 2020. Front Immunol. 2021. PMID: 33552080 Free PMC article. Review.
Cited by
- Intermolecular interactions in a 44 kDa interferon-receptor complex detected by asymmetric reverse-protonation and two-dimensional NOESY.
Nudelman I, Akabayov SR, Schnur E, Biron Z, Levy R, Xu Y, Yang D, Anglister J. Nudelman I, et al. Biochemistry. 2010 Jun 29;49(25):5117-33. doi: 10.1021/bi100041f. Biochemistry. 2010. PMID: 20496919 Free PMC article. - Site-selective protein conjugation at histidine.
Peciak K, Laurine E, Tommasi R, Choi JW, Brocchini S. Peciak K, et al. Chem Sci. 2018 Oct 9;10(2):427-439. doi: 10.1039/c8sc03355b. eCollection 2019 Jan 14. Chem Sci. 2018. PMID: 30809337 Free PMC article. - Type-1 interferons contribute to oxygen glucose deprivation induced neuro-inflammation in BE(2)M17 human neuroblastoma cells.
Minter MR, Zhang M, Ates RC, Taylor JM, Crack PJ. Minter MR, et al. J Neuroinflammation. 2014 Mar 6;11:43. doi: 10.1186/1742-2094-11-43. J Neuroinflammation. 2014. PMID: 24602263 Free PMC article. - Differential levels of IFNα subtypes in autoimmunity and viral infection.
Bondet V, Rodero MP, Posseme C, Bost P, Decalf J, Haljasmägi L, Bekaddour N, Rice GI, Upasani V, Herbeuval JP, Reynolds JA, Briggs TA, Bruce IN, Mauri C, Isenberg D, Menon M, Hunt D, Schwikowski B, Mariette X, Pol S, Rozenberg F, Cantaert T, Eric Gottenberg J, Kisand K, Duffy D. Bondet V, et al. Cytokine. 2021 Aug;144:155533. doi: 10.1016/j.cyto.2021.155533. Epub 2021 Apr 30. Cytokine. 2021. PMID: 33941444 Free PMC article. - Structural and dynamic determinants of type I interferon receptor assembly and their functional interpretation.
Piehler J, Thomas C, Garcia KC, Schreiber G. Piehler J, et al. Immunol Rev. 2012 Nov;250(1):317-34. doi: 10.1111/imr.12001. Immunol Rev. 2012. PMID: 23046138 Free PMC article. Review.
References
- Pestka S, Krause CD, Walter MR. Interferons, interferon-like cytokines, and their receptors. Immunol Rev. 2004;202:8–32. - PubMed
- Novick D, Cohen B, Rubinstein M. The human interferon alpha/beta receptor: characterization and molecular cloning. Cell. 1994;77:391–400. - PubMed
- Uze G, Lutfalla G, Mogensen KE. Alpha and beta interferons and their receptor and their friends and relations. J Interferon Cytokine Res. 1995;15:3–26. - PubMed
- Piehler J, Schreiber G. Mutational and structural analysis of the binding interface between type I interferons and their receptor Ifnar2. J Mol Biol. 1999;294:223–237. - PubMed
- Lamken P, Gavutis M, Peters I, Van der Heyden J, Uze G, Piehler J. Functional cartography of the ectodomain of the type I interferon receptor subunit ifnar1. J Mol Biol. 2005;350:476–488. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources