PHD fingers: epigenetic effectors and potential drug targets - PubMed (original) (raw)
Review
PHD fingers: epigenetic effectors and potential drug targets
Catherine A Musselman et al. Mol Interv. 2009 Dec.
Abstract
The plant homeodomain (PHD) finger is found in many chromatin-remodeling proteins. This small approximately 65-residue domain functions as an "effector" that binds specific epigenetic marks on histone tails, recruiting transcription factors and nucleosome-associated complexes to chromatin. Mutations in the PHD finger or deletion of this domain are linked to a number of human diseases, including cancer, mental retardation, and immunodeficiency. PHD finger-containing proteins may become valuable diagnostic markers and targets to prevent and treat these disorders. In this review, we highlight the progress recently made in understanding the functional significance of chromatin targeting by mammalian PHD fingers, detail the molecular mechanisms and structural features of "histone code" recognition, and discuss the therapeutic potential of PHD fingers.
Figures
Tatiana G. Kutateladze, PhD, is an Associate Professor of Pharmacology at the University of Colorado Denver School of Medicine. Epigenetic gene regulation and phosphoinositide (PI) signaling are her two major areas of interest. The research in her laboratory is primarily focused on the study of the atomic-resolution structures and functions of proteins involved in recognition of epigenetic marks and phosphorylated PIs. E-mail Tatiana.
; fax 303-724-3663.
Catherine A. Musselman, PhD, graduated from the University of Michigan in 2007. She is currently a postdoctoral fellow in Tatiana Kutateladze’s laboratory in the Department of Pharmacology at the University of Colorado Denver School of Medicine. She investigates the molecular mechanisms of histone binding by PHD fingers and other epigenetic reader modules. E-mail Catherine.
; fax 303-724-3663.
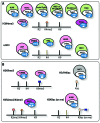
Figure 1
PHD fingers as epigenetic effectors. Shown are the PHD fingers and associated complexes that recognize A) H3K4me3 (orange triangle) or unH3 (yellow bar) and B) other PTMs (including H3 methylated at both Arg2 (R2me2, salmon hexagon) and Lys4 (K4me3, orange triangle) and H3K9me3 or H3K9ac (taupe oval or blue rectangle). Complexes are colored by their function as promoters of gene activation (green), repression (pink), or recombination (purple).
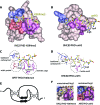
Figure 2
Structural bases of histone code interpretation by PHD fingers. The crystal structures of (A) H3K4me3-bound ING2 PHD and (B) unH3-bound BHC80 PHD. The binding pockets for Ala1, Arg2, and Lys4me3/Lys4 are colored blue, salmon, and purple, respectively. The detailed coordination of these residues is highlighted (dashed circles) for (C) BPTF PHD-H3K4me3 and (D) BHC80 PHD-unH3. E) The PHD finger fold is characterized by the canonical Cys4-His-Cys3 (C4HC3) sequence that coordinates two zinc ions and contributes to the typical two-stranded anti-parallel β sheet structure. F) The dichotomy of coordination of Arg2 in the RAG2 PHD-H3K4me3 complex (left) and AIRE PHD-unH3 complex (right).
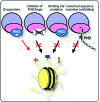
Figure 3
PHD fingers in disease. Dysregulation of the PHD finger activities can occur as a result of its deletion, from mutations in the histone binding site, or mutations in the canonical sequence leading to misfolding. The PHD finger may also promote disease by deleterious recruitment of oncoproteins to chromatin.
References
- Schindler U, Beckmann, H, Cashmore, AR and Schindler, TF. HAT3.1, a novel Arabidopsis homeodomain protein containing a conserved cysteine-rich region. Plant J. 4, 137–150 (1993). - PubMed
- Luger K, Mader, AW, Richmond, RK, Sargent, DF and Richmond, TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. [see comment]. Nature 389, 251–260 (1997). - PubMed
- Kouzarides T. Chromatin modifications and their function. Cell 128, 693–705 (2007). - PubMed
- Berger SL. The complex language of chromatin regulation during transcription. Nature 447, 407–412 (2007). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical