Nanostructural and transcriptomic analyses of human saliva derived exosomes - PubMed (original) (raw)
Nanostructural and transcriptomic analyses of human saliva derived exosomes
Viswanathan Palanisamy et al. PLoS One. 2010.
Abstract
Background: Exosomes, derived from endocytic membrane vesicles are thought to participate in cell-cell communication and protein and RNA delivery. They are ubiquitous in most body fluids (breast milk, saliva, blood, urine, malignant ascites, amniotic, bronchoalveolar lavage, and synovial fluids). In particular, exosomes secreted in human saliva contain proteins and nucleic acids that could be exploited for diagnostic purposes. To investigate this potential use, we isolated exosomes from human saliva and characterized their structural and transcriptome contents.
Methodology: Exosomes were purified by differential ultracentrifugation and identified by immunoelectron microscopy (EM), flow cytometry, and Western blot with CD63 and Alix antibodies. We then described the morphology, shape, size distribution, and density using atomic force microscopy (AFM). Microarray analysis revealed that 509 mRNA core transcripts are relatively stable and present in the exosomes. Exosomal mRNA stability was determined by detergent lysis with RNase A treatment. In vitro, fluorescently labeled saliva exosomes could communicate with human keratinocytes, transferring their genetic information to human oral keratinocytes to alter gene expression at a new location.
Conclusion: Our findings are consistent with the hypothesis that exosomes shuttle RNA between cells and that the RNAs present in the exosomes may be a possible resource for disease diagnostics.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
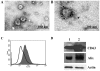
Figure 1. EM image of human saliva showing round-shaped exosomes.
The 120,000×g pellets from saliva were used for exosomes analysis. (A) Electron micrographs of saliva exosomes were fixed in 2% formaldehyde and contrasted using 2% uranyl acetate. The image shows small vesicles of ∼60 nm in diameter. (B) Exosomes were labeled with immunogold anti-CD63. Note the immunoreactivity of CD63 on the surface of the single exosome. (C) Representative FACS analysis of exosomes showing expression of CD63. Open trace shows the saliva exosomes, filled trace shows saliva exosomes incubated with latex beads and stained with anti-CD63 followed by secondary Alexa Fluor 488-conjugated antibody. (D) Western blot analysis of saliva exosomes using an antibody against CD63 and Alix. Lane 1 is the protein extract of normal saliva, and lane 2 contains protein from the exosome pellet obtained from ultracentrifugation.
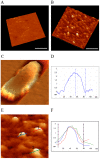
Figure 2. AFM images of saliva exosomes.
Exosomes (panels B_–_F) were adsorbed to WGA-coated mica surfaces. (A) Topography images were obtained with the use of the Mac mode in water (negative control—no exosomes). (B) A 3D AFM image of isolated exosomes adhering to a mica sheet. The bar denotes 200µM. (C) A high-resolution single image of the exosome structure on the mica. (D) Graphical representation of height and width of a single exosome. (E) Size distribution of several saliva exosomes imaged with AFM. (F) Graphical representation of the size distribution of exosomes showing near homogeneity with respect to height and width.
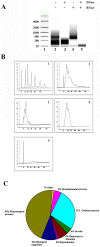
Figure 3. Exosomes contain mRNA species.
(A) RNA from saliva exosomes was detected using an Agilent bioanalyser. Lane 1, RNA ladder showing sizes of the nucleotides on the left. Representative lanes (2–5) showing sizes of the mRNA species identified using an Agilent bioanalyser electrophorogram. The saliva exosomal RNA contains no ribosomal RNA as seen by the small heterogeneous RNA fragments (<200 nucleotides). (B) Bioanalyzer graphical data shows the size distribution of total RNA extracted from saliva exosomes (1) profile of RNA standard (2) total RNA extracted from saliva exosomes without any treatments (3) total RNA treated with DNase (4) total RNA treated with RNase A and (5) total RNA treated with both DNase and RNase A. (C) The biological process ontology of the 509 core mRNA species identified in the saliva exosomes.
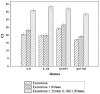
Figure 4. Saliva exosomes treated with Triton X-100 and RNase had different RNA content compared to control, indicating that RNA is protected inside the exosomes.
Higher Ct values represent lower RNA content. Error bars denote SEM (n = 3).
Figure 5. Oral keratinocytes (5×107 cells/well) were incubated for 24 hr in KSFM media with fluorescently labeled exosomes and examined under fluorescence microscopy.
The lysed lanes serve as a negative control. Magnification was 10×, and the smaller boxed panels represent magnification of 40×. Note the fluorescence intensity increases with increasing amounts of exosomes (32 and 64 µl, respectively).
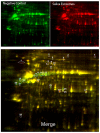
Figure 6. Identification of differentially expressed proteins from oral keratinocytes treated with saliva exosomes by 2-DIGE.
Proteins from cells treated with the negative control were labeled with Cy3 (green) and proteins from cells treated with saliva exosomes were labeled with Cy5 (red). Isoelectric focusing was carried out at pH 3–10, and 2D separation was performed with 8–14% gradient SDS-PAGE. The negative control represents protein profiles of keratinocytes treated with detergent lysed exosomes. The bottom gel image reveals differentially expressed proteins in the control and treated samples after merging. Protein spots shown in red are presumably due to upregulation by exosome treatment, and those in green are due to downregulation by exosome treatment. Such spots are circled and numbered.
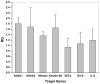
Figure 7. Protein expression modulated in saliva exosomes.
ANXA1, ANXA2, moesin, keratin-6A, EEF2, OS-9, and IL-8 RNA expression was normalized using a factor calculated from β-Actin gene expression.
Similar articles
- Human saliva, plasma and breast milk exosomes contain RNA: uptake by macrophages.
Lässer C, Alikhani VS, Ekström K, Eldh M, Paredes PT, Bossios A, Sjöstrand M, Gabrielsson S, Lötvall J, Valadi H. Lässer C, et al. J Transl Med. 2011 Jan 14;9:9. doi: 10.1186/1479-5876-9-9. J Transl Med. 2011. PMID: 21235781 Free PMC article. - RNA-containing exosomes in human nasal secretions.
Lässer C, O'Neil SE, Ekerljung L, Ekström K, Sjöstrand M, Lötvall J. Lässer C, et al. Am J Rhinol Allergy. 2011 Mar-Apr;25(2):89-93. doi: 10.2500/ajra.2011.25.3573. Epub 2010 Dec 17. Am J Rhinol Allergy. 2011. PMID: 21172122 - Body fluid derived exosomes as a novel template for clinical diagnostics.
Keller S, Ridinger J, Rupp AK, Janssen JW, Altevogt P. Keller S, et al. J Transl Med. 2011 Jun 8;9:86. doi: 10.1186/1479-5876-9-86. J Transl Med. 2011. PMID: 21651777 Free PMC article. - Exosomes: proteomic insights and diagnostic potential.
Simpson RJ, Lim JW, Moritz RL, Mathivanan S. Simpson RJ, et al. Expert Rev Proteomics. 2009 Jun;6(3):267-83. doi: 10.1586/epr.09.17. Expert Rev Proteomics. 2009. PMID: 19489699 Review. - Ascent of atomic force microscopy as a nanoanalytical tool for exosomes and other extracellular vesicles.
Sharma S, LeClaire M, Gimzewski JK. Sharma S, et al. Nanotechnology. 2018 Apr 3;29(13):132001. doi: 10.1088/1361-6528/aaab06. Nanotechnology. 2018. PMID: 29376505 Review.
Cited by
- Exosomes: Implications in HIV-1 Pathogenesis.
Madison MN, Okeoma CM. Madison MN, et al. Viruses. 2015 Jul 20;7(7):4093-118. doi: 10.3390/v7072810. Viruses. 2015. PMID: 26205405 Free PMC article. Review. - Characterization of human plasma-derived exosomal RNAs by deep sequencing.
Huang X, Yuan T, Tschannen M, Sun Z, Jacob H, Du M, Liang M, Dittmar RL, Liu Y, Liang M, Kohli M, Thibodeau SN, Boardman L, Wang L. Huang X, et al. BMC Genomics. 2013 May 10;14:319. doi: 10.1186/1471-2164-14-319. BMC Genomics. 2013. PMID: 23663360 Free PMC article. - Mast Cell-Derived Exosomes Promote Th2 Cell Differentiation via OX40L-OX40 Ligation.
Li F, Wang Y, Lin L, Wang J, Xiao H, Li J, Peng X, Dai H, Li L. Li F, et al. J Immunol Res. 2016;2016:3623898. doi: 10.1155/2016/3623898. Epub 2016 Mar 15. J Immunol Res. 2016. PMID: 27066504 Free PMC article. - Salivary exosomes: properties, medical applications, and isolation methods.
Cheshmi B, Cheshomi H. Cheshmi B, et al. Mol Biol Rep. 2020 Aug;47(8):6295-6307. doi: 10.1007/s11033-020-05659-1. Epub 2020 Jul 16. Mol Biol Rep. 2020. PMID: 32676813 Review. - Non-coding RNAs in saliva: emerging biomarkers for molecular diagnostics.
Majem B, Rigau M, Reventós J, Wong DT. Majem B, et al. Int J Mol Sci. 2015 Apr 17;16(4):8676-98. doi: 10.3390/ijms16048676. Int J Mol Sci. 2015. PMID: 25898412 Free PMC article. Review.
References
- Delcayre A, Estelles A, Sperinde J, Roulon T, Paz P, et al. Exosome Display technology: applications to the development of new diagnostics and therapeutics. Blood Cells Mol Dis. 2005;35:158–168. - PubMed
- Heijnen HF, Schiel AE, Fijnheer R, Geuze HJ, Sixma JJ. Activated platelets release two types of membrane vesicles: microvesicles by surface shedding and exosomes derived from exocytosis of multivesicular bodies and alpha-granules. Blood. 1999;94:3791–3799. - PubMed
- Rozmyslowicz T, Majka M, Kijowski J, Murphy SL, Conover DO, et al. Platelet- and megakaryocyte-derived microparticles transfer CXCR4 receptor to CXCR4-null cells and make them susceptible to infection by X4-HIV. AIDS. 2003;17:33–42. - PubMed
- Blanchard N, Lankar D, Faure F, Regnault A, Dumont C, et al. TCR activation of human T cells induces the production of exosomes bearing the TCR/CD3/zeta complex. J Immunol. 2002;168:3235–3241. - PubMed
Publication types
MeSH terms
Grants and funding
- K99 DE018165/DE/NIDCR NIH HHS/United States
- U01 DE016275/DE/NIDCR NIH HHS/United States
- P20RR017696/RR/NCRR NIH HHS/United States
- U01DE16275/DE/NIDCR NIH HHS/United States
- R00DE018165/DE/NIDCR NIH HHS/United States
- R01DE017170/DE/NIDCR NIH HHS/United States
- P20 RR017696/RR/NCRR NIH HHS/United States
- R00 DE018165/DE/NIDCR NIH HHS/United States
- R01 DE017170/DE/NIDCR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous