MolProbity: all-atom structure validation for macromolecular crystallography - PubMed (original) (raw)
MolProbity: all-atom structure validation for macromolecular crystallography
Vincent B Chen et al. Acta Crystallogr D Biol Crystallogr. 2010 Jan.
Abstract
MolProbity is a structure-validation web service that provides broad-spectrum solidly based evaluation of model quality at both the global and local levels for both proteins and nucleic acids. It relies heavily on the power and sensitivity provided by optimized hydrogen placement and all-atom contact analysis, complemented by updated versions of covalent-geometry and torsion-angle criteria. Some of the local corrections can be performed automatically in MolProbity and all of the diagnostics are presented in chart and graphical forms that help guide manual rebuilding. X-ray crystallography provides a wealth of biologically important molecular data in the form of atomic three-dimensional structures of proteins, nucleic acids and increasingly large complexes in multiple forms and states. Advances in automation, in everything from crystallization to data collection to phasing to model building to refinement, have made solving a structure using crystallography easier than ever. However, despite these improvements, local errors that can affect biological interpretation are widespread at low resolution and even high-resolution structures nearly all contain at least a few local errors such as Ramachandran outliers, flipped branched protein side chains and incorrect sugar puckers. It is critical both for the crystallographer and for the end user that there are easy and reliable methods to diagnose and correct these sorts of errors in structures. MolProbity is the authors' contribution to helping solve this problem and this article reviews its general capabilities, reports on recent enhancements and usage, and presents evidence that the resulting improvements are now beneficially affecting the global database.
Figures
Figure 1
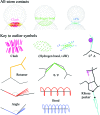
An outlier legend, showing each symbol used in a MolProbity multi-criterion kinemage and illustrating the relationship of the three types of all-atom contact to the atomic van der Waals (vdW) surfaces (spheres of small gray dots). The symbols for favorable hydrogen bonds and vdW contacts are included for completeness, as well as the hot-pink spikes of a clash outlier. A Cβ deviation of ≥0.25 Å is shown as a magenta ball centered on the ideal Cβ position and tangent to the modeled position. Bad rotamers are shown as gold side chains and Ramachandran outliers as heavy green lines to the midpoints of the two peptides. Bond-angle outliers are indicated by a fan of lines from the ideal to the modeled bond (red if wide, blue if narrow). Bond-length outliers are indicated as stretched (red) or compressed (blue) springs. A suspicious ribose pucker is diagnosed by the perpendicular distance from the 3′ (following) phosphate to the line of the glycosidic C1′—N1/9 bond and is flagged by a representation of that construction (in magenta if too short, as here, and in purple if too long).
Figure 2
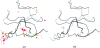
Two multi-criterion validation kinemages illustrating the successful outcome of an overall process of MolProbity diagnosis and structure improvement. (a) The original
1lpl
Cap-Gly structure (Li et al., 2002 ▶) shows three major clusters of clash, rotamer and Ramachandran problems plus a few isolated outliers. (b) The corrected
1tov
structure (Arendall et al., 2005 ▶) has essentially no outliers, a 4% lower R free, a bound sulfate and an additional turn of helix at the N-terminus.
Figure 3
The simple ‘flip’ correction of a Gln side-chain amide in the
2dq4
threonine 3-dehydrogenase structure (R. Omi, T. Yao, M. Goto, I. Miyahara & K. Hirotsu, unpublished work), a better-than-average 2.5 Å resolution structure. Both orientations make a hydrogen bond to the crystallographic water, but the original has a serious internal clash of the NH2 group with its own Cβ hydrogen.
Figure 4
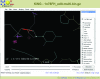
A MolProbity results summary and sortable multi-criterion chart for the
1n78
Glu tRNA-synthetase complex at 2.1 Å resolution (Sekine et al., 2003 ▶). The summary gives numerical values, goals and relative percentiles for clashscore, torsion angle and geometry criteria for both protein and nucleic acid components, with traffic light color-coding for good and bad values. Below the summary is a short extract from the detailed chart with values and specifics for each criterion on each residue. Notice that outliers (highlighted) tend to cluster.
Figure 5
The general case Ramachandran kinemage and the Cβ deviation kinemage for file
2dq4
. In (a) the ϕ, ψ values for each residue are plotted on a background of the smoothed contours from high-quality data (see text). Over 98% lie inside the inner ‘favored’ 98% contour, but there are seven outliers outside the outer 99.95% contour. Gly, Pro and pre-Pro residues are on separate plots (not shown). In (b) the Cβ deviation kinemage shows each residue’s Cβ position relative to an ideal Cβ and its three bond vectors (gray lines). Circles mark the deviation distances, with the yellow circle at the 0.25 Å cutoff for outliers. Most of the distribution is good, but an adjacent Leu and Trp in each chain (labeled) are part of an outlier cluster and probably reflect distortions caused by a local fitting problem.
Figure 6
Close-up of a ribose-pucker outlier in the multi-criterion kinemage for
1n78
, with backbone and bases turned on. C574 has a short phosphate-to-glycosidic bond perpendicular (magenta line and cross), but was fitted with an intermediate pucker near C3′-endo. The bad pucker torques the connected groups strongly, probably causing the bond-angle outliers (red) and steric clashes (hot-pink spikes). Note that C574 is in the binding interface between RNA (white backbone) and enzyme (yellow backbone) close to the active site.
Figure 7
Rebuilding of a backward-fitted Leu side chain in KiNG off-line in the DNA polymerase
1xwl
at 1.7 Å resolution (Kiefer et al., 1997 ▶). The original (left) fits the density fairly well but is a rotamer outlier with a clash. One of the two best Leu rotamers also fits the density well with good all-atom packing. The top view (right) shows the 180° relationship of the two conformations.
Figure 8
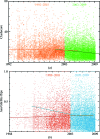
_MolProbity_-relevant quality criteria as a function of time for all structures in the PDB at a middle range of resolution, separately fitted before and after introduction of the web site. (a) All-atom clashscore (see §2.2); (b) percentage of Asn/Gln/His flips (see §2.1).
Similar articles
- MolProbity: all-atom contacts and structure validation for proteins and nucleic acids.
Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB 3rd, Snoeyink J, Richardson JS, Richardson DC. Davis IW, et al. Nucleic Acids Res. 2007 Jul;35(Web Server issue):W375-83. doi: 10.1093/nar/gkm216. Epub 2007 Apr 22. Nucleic Acids Res. 2007. PMID: 17452350 Free PMC article. - MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes.
Davis IW, Murray LW, Richardson JS, Richardson DC. Davis IW, et al. Nucleic Acids Res. 2004 Jul 1;32(Web Server issue):W615-9. doi: 10.1093/nar/gkh398. Nucleic Acids Res. 2004. PMID: 15215462 Free PMC article. - New tools in MolProbity validation: CaBLAM for CryoEM backbone, UnDowser to rethink "waters," and NGL Viewer to recapture online 3D graphics.
Prisant MG, Williams CJ, Chen VB, Richardson JS, Richardson DC. Prisant MG, et al. Protein Sci. 2020 Jan;29(1):315-329. doi: 10.1002/pro.3786. Epub 2019 Dec 10. Protein Sci. 2020. PMID: 31724275 Free PMC article. - Nucleic acid X-ray crystallography via direct selenium derivatization.
Lin L, Sheng J, Huang Z. Lin L, et al. Chem Soc Rev. 2011 Sep;40(9):4591-602. doi: 10.1039/c1cs15020k. Epub 2011 Jun 13. Chem Soc Rev. 2011. PMID: 21666919 Review. - Collection of X-Ray Diffraction Data from Macromolecular Crystals.
Dauter Z. Dauter Z. Methods Mol Biol. 2017;1607:165-184. doi: 10.1007/978-1-4939-7000-1_7. Methods Mol Biol. 2017. PMID: 28573573 Free PMC article. Review.
Cited by
- Human transforming growth factor β type I receptor in complex with kinase inhibitor SB505124.
Rodriguez Buitrago JA, Landström M, Wolf-Watz M. Rodriguez Buitrago JA, et al. Acta Crystallogr F Struct Biol Commun. 2024 Nov 1;80(Pt 11):314-319. doi: 10.1107/S2053230X24010094. Epub 2024 Oct 23. Acta Crystallogr F Struct Biol Commun. 2024. PMID: 39441620 Free PMC article. - Infectious parvovirus B19 circulates in the blood coated with active host protease inhibitors.
Lee H, Assaraf R, Subramanian S, Goetschius D, Bieri J, DiNunno NM, Leisi R, Bator CM, Hafenstein SL, Ros C. Lee H, et al. Nat Commun. 2024 Nov 5;15(1):9543. doi: 10.1038/s41467-024-53794-1. Nat Commun. 2024. PMID: 39500886 Free PMC article. - Insights into the structure of mature streptavidin C1 from Streptomyces cinnamonensis reveal the self-binding of the extension C-terminal peptide to biotin-binding sites.
Jeon BJ, Kim S, Kim MS, Lee JH, Kim BS, Hwang KY. Jeon BJ, et al. IUCrJ. 2021 Jan 11;8(Pt 2):168-177. doi: 10.1107/S2052252520015675. eCollection 2021 Mar 1. IUCrJ. 2021. PMID: 33708394 Free PMC article. - Structure of a Ca(2+)/CaM:Kv7.4 (KCNQ4) B-helix complex provides insight into M current modulation.
Xu Q, Chang A, Tolia A, Minor DL Jr. Xu Q, et al. J Mol Biol. 2013 Jan 23;425(2):378-94. doi: 10.1016/j.jmb.2012.11.023. Epub 2012 Nov 23. J Mol Biol. 2013. PMID: 23178170 Free PMC article. - The UmuC subunit of the E. coli DNA polymerase V shows a unique interaction with the β-clamp processivity factor.
Patoli AA, Winter JA, Bunting KA. Patoli AA, et al. BMC Struct Biol. 2013 Jul 4;13:12. doi: 10.1186/1472-6807-13-12. BMC Struct Biol. 2013. PMID: 23822808 Free PMC article.
References
- Adams, P. D., Grosse-Kunstleve, R. W., Hung, L.-W., Ioerger, T. R., McCoy, A. J., Moriarty, N. W., Read, R. J., Sacchettini, J. C., Sauter, N. K. & Terwilliger, T. C. (2002). Acta Cryst. D58, 1948–1954. - PubMed
- Arendall, W. B. III, Tempel, W., Richardson, J. S., Zhou, W., Wang, S., Davis, I. W., Liu, Z.-J., Rose, J. P., Carson, W. M., Luo, M., Richardson, D. C. & Wang, B.-C. (2005). J. Struct. Funct. Genomics, 6, 1–11. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases