Allosteric regulation of Argonaute proteins by miRNAs - PubMed (original) (raw)
Allosteric regulation of Argonaute proteins by miRNAs
Sergej Djuranovic et al. Nat Struct Mol Biol. 2010 Feb.
Abstract
Small interfering RNAs (siRNAs) and microRNAs (miRNAs) bind to Argonaute (AGO) family proteins to form a related set of effector complexes that have diverse roles in post-transcriptional gene regulation throughout the eukaryotic lineage. Here sequence and structural analysis of the MID domain of the AGO proteins identified similarities with a family of allosterically regulated bacterial ligand-binding domains. We used in vitro and in vivo approaches to show that certain AGO proteins (those involved in translational repression) have conserved this functional allostery between two distinct sites, one involved in binding miRNA-target duplex and the other in binding the 5' cap feature (m(7)GpppG) of eukaryotic mRNAs. This allostery provides an explanation for how miRNA-bound effector complexes may avoid indiscriminate repressive action (mediated through binding interactions with the cap) before full target recognition.
Figures
Figure 1
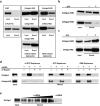
Bioinformatic analysis of Argonaute proteins. a, Cartoon representation of the T. Thermophilus Argonaute (PDB: 3dlb) showing the N-terminal domain in cyan, PAZ domain in blue, MID domain in red and PIWI domain in magenta. b, Superimposition of the modeled DmAgo1 MID domain (red) with similar structures: N-domain of sugar transporter from C. phytofermentans (green, PDB: 3brs, residues 5-98 and 246-263), ligand binding domain of E. coli PurR purine repressor (blue, PDB: 2pua, residues 60-147 and 294-311) and ligand-sensitive negative regulator from P. aeruginosa, AmiC (orange, PDB: 1qo0, residues 127-232). c, Structure of the M. musculus eIF4E bound to m7GDP (left, PDB: 1ej1, m7GDP in blue) and modeled DmAgo1 MID domain (right). Tryptophan residues of eIF4E (W56 and W102), involved in binding of m7GDP, and “equivalent” phenylalanine residues (F560 and F595) of modeled DmAgo1 MID domain are shown in orange. d, Cluster analysis of MID-domain sequences from Argonaute/PIWI protein family. Each sequence is represented by one dot. Darker lines represent pairwise connections with very low P-values and lighter ones those with higher values, closer to the cutoff (10−10). Positions of protein sequences discussed in this study are labeled with colored dots.
Figure 2
Binding of Argonaute proteins to m7GTP-Sepharose reveal allosteric behavior. a, Binding of purified Argonaute MID-domains and PurR protein to the m7GTP-Sepharose (Dm, D. melanogaster; Ce, C. elegans; Hs, H. sapiens; Ec, E. coli). miRNA and siRNA related Argonautes are shown in separate boxes. MBP is used as a negative control. b, Binding of DmAgo1-MID and DmAgo2-MID domains to m7GTP-Sepharose in the presence of GTP. The total amount of nucleotide is 500 μM, input (10%) and bound fraction (40%) were analyzed by western blotting. c, Binding of DmAgo1 and CeAlg1 MID domains to m7GTP-Sepharose is stimulated by addition of 250 μM GTP. Input (10%) and bound fraction (50%) were analyzed by western blotting. d, Binding of full-length DmAgo1, and not of DmAgo2ΔQ, to m7GTP-Sepharose is specific and stimulated by addition of short RNA (23-mer). Binding to m7GTP-resin is competed by addition of m7GpppG. Binding of both, DmAgo1 and DmAgo2ΔQ, to GTP- and GMP-Sepharose is competed by addition of short RNA (23-mer). m7GpppG shows no effect on binding to these resins. Input (20%) and bound fraction (25%) were analyzed by western blotting. e, Binding of full-length DmAgo1 to m7GTP-Sepharose in presence of increasing amounts of short RNA (23-mer). Input (20%) and bound fraction (20%) in assays were analyzed by western blotting using polyclonal anti-MBP (NEB) antibody.
Figure 3
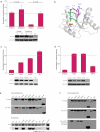
Filter binding assays indicate two allosterically regulated nucleotide binding sites. Binding of 32P-labeled bantam to DmAgo1 in a and DmAgo2ΔQ in b in the presence of non-labeled RNAs of varying lengths (5, 15, 23 and 30 nts). Competition between labeled and non-labeled RNAs was performed by titration of the non-labeled species and represented by the ratio of bound 32P-bantam in the presence and absence of competing RNA species. Influence of different nucleotide substrates on the 32P-bantam binding to DmAgo1 in c and DmAgo2ΔQ in d. Nucleotide substrates were titrated and the amount of the bound 32P-bantam was followed. Values represent the ratio of bound 32P-bantam in the presence and absence of indicated nucleotide substrate. e, Binding of bantam duplex (miRNA*) to DmAgo1 and DmAgo2ΔQ stimulates only binding of DmAgo1 to 3H-labeled m7GpppG mRNA. f, Influence of different nucleotide substrates on binding of 3H-m7GpppG mRNA to DmAgo1. Error bars represent the standard error from at least three experiments.
Figure 4
Mutational analysis of Argonaute proteins in Drosophila S2 cells. a, c, d, S2 cells were transfected with a mixture of three plasmids: either the F-Luc-0BoxB or -5BoxB reporter, R-Luc transfection control and the indicated Argonaute encoding construct. HA-tagged Argonaute proteins were either fused with λN sequences or not, as indicated. For each DmAgo variant, F-Luc values were normalized to R-Luc and triplicates were averaged. Tagged (+N) values were normalized to untagged (−N) values. Western blots show expression of each tested variant. Error bars represent the standard deviation from at least three independent experiments. a, Repression of the FLuc reporter by DmAgo1 (but not DmAgo2ΔQ) in the tethering assay depends on the presence of 5 boxB stem loops in the reporter 3’UTR. b, Cartoon representation of modeled DmAgo1 MID domain with sites of mutation and 5’ nucleotide of bound miRNA indicated (5’-U in pink). Control surface residues K615 and K640 are shown in dark blue while buried residues, F560 and F595, are shown in light blue. c, Mutations in the 5’-end miRNA binding site of DmAgo1. d, Mutation of residues 627-630 in loop 3 of DmAgo1. e, f, S2 cells were transfected with indicated HA-tagged constructs; null contained no transfected protein. e, MicroRNA binding by DmAgo1 (and variants) evaluated by HA-pull-down, and subsequent northern analysis probing for bantam in precipitated samples. “bantam” lane contains 1 pmol of bantam miRNA (Dharmacon). Quantitation of northern blots indicates extent of binding differences. f, Binding to m7GTP-Sepharose by HA-DmAgo1 (and variants).
Figure 5
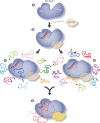
Cartoon describing potential role of Argonaute allostery in promoting translational repression. a, Argonaute protein with various domains indicated. b, Initial interaction between Argonaute, miRNA and potentially GW182 (transparent) are shown. Next, miRNA-bound Argonaute either c, samples messages independent of cap binding or d, specifically scans capped messages, both pathways leading to e, a fully engaged Argonaute protein ready for translational repression. miRNA:target duplex, cap and GW182 are all likely bound in this final stage.
Similar articles
- Alternative RISC assembly: binding and repression of microRNA-mRNA duplexes by human Ago proteins.
Janas MM, Wang B, Harris AS, Aguiar M, Shaffer JM, Subrahmanyam YV, Behlke MA, Wucherpfennig KW, Gygi SP, Gagnon E, Novina CD. Janas MM, et al. RNA. 2012 Nov;18(11):2041-55. doi: 10.1261/rna.035675.112. Epub 2012 Sep 27. RNA. 2012. PMID: 23019594 Free PMC article. - Effects of the PIWI/MID domain of Argonaute protein on the association of miRNAi's seed base with the target.
Wang Z, Wang Y, Liu T, Wang Y, Zhang W. Wang Z, et al. RNA. 2019 May;25(5):620-629. doi: 10.1261/rna.069328.118. Epub 2019 Feb 15. RNA. 2019. PMID: 30770397 Free PMC article. - Artificial tethering of Argonaute proteins for studying their role in translational repression of target mRNAs.
Eckhardt S, Szostak E, Yang Z, Pillai R. Eckhardt S, et al. Methods Mol Biol. 2011;725:191-206. doi: 10.1007/978-1-61779-046-1_13. Methods Mol Biol. 2011. PMID: 21528455 - A parsimonious model for gene regulation by miRNAs.
Djuranovic S, Nahvi A, Green R. Djuranovic S, et al. Science. 2011 Feb 4;331(6017):550-3. doi: 10.1126/science.1191138. Science. 2011. PMID: 21292970 Free PMC article. Review. - MicroRNAs repress translation of m7Gppp-capped target mRNAs in vitro by inhibiting initiation and promoting deadenylation.
Standart N, Jackson RJ. Standart N, et al. Genes Dev. 2007 Aug 15;21(16):1975-82. doi: 10.1101/gad.1591507. Genes Dev. 2007. PMID: 17699746 Review. No abstract available.
Cited by
- Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state.
Le Thomas A, Rogers AK, Webster A, Marinov GK, Liao SE, Perkins EM, Hur JK, Aravin AA, Tóth KF. Le Thomas A, et al. Genes Dev. 2013 Feb 15;27(4):390-9. doi: 10.1101/gad.209841.112. Epub 2013 Feb 7. Genes Dev. 2013. PMID: 23392610 Free PMC article. - Temporospatial guidance of activity-dependent gene expression by microRNA: mechanisms and functional implications for neural plasticity.
Kiltschewskij D, Cairns MJ. Kiltschewskij D, et al. Nucleic Acids Res. 2019 Jan 25;47(2):533-545. doi: 10.1093/nar/gky1235. Nucleic Acids Res. 2019. PMID: 30535081 Free PMC article. Review. - Argonaute protein as a linker to command center of physiological processes.
Wei K, Wu L, Chen Y, Lin Y, Wang Y, Liu X, Xie D. Wei K, et al. Chin J Cancer Res. 2013 Aug;25(4):430-41. doi: 10.3978/j.issn.1000-9604.2013.08.13. Chin J Cancer Res. 2013. PMID: 23997530 Free PMC article. - Drosophila Argonaute 1 and its miRNA biogenesis partners are required for oocyte formation and germline cell division.
Azzam G, Smibert P, Lai EC, Liu JL. Azzam G, et al. Dev Biol. 2012 May 15;365(2):384-94. doi: 10.1016/j.ydbio.2012.03.005. Epub 2012 Mar 14. Dev Biol. 2012. PMID: 22445511 Free PMC article. - Co-option of the piRNA pathway to regulate neural crest specification.
Galton R, Fejes-Toth K, Bronner ME. Galton R, et al. Sci Adv. 2022 Aug 12;8(32):eabn1441. doi: 10.1126/sciadv.abn1441. Epub 2022 Aug 10. Sci Adv. 2022. PMID: 35947657 Free PMC article.
References
- Wu L, Belasco JG. Let me count the ways: mechanisms of gene regulation by miRNAs and siRNAs. Mol Cell. 2008;29:1–7. - PubMed
- Kiriakidou M, et al. An mRNA m(7)G cap binding-like motif within human Ago2 represses translation. Cell. 2007;129:1141–1151. - PubMed
- Pillai RS, et al. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–6. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous