The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus - PubMed (original) (raw)
. 2009 Dec 24;139(7):1243-54.
doi: 10.1016/j.cell.2009.12.017.
I-Chueh Huang, Yair Benita, Sinu P John, Manoj N Krishnan, Eric M Feeley, Bethany J Ryan, Jessica L Weyer, Louise van der Weyden, Erol Fikrig, David J Adams, Ramnik J Xavier, Michael Farzan, Stephen J Elledge
Affiliations
- PMID: 20064371
- PMCID: PMC2824905
- DOI: 10.1016/j.cell.2009.12.017
The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus
Abraham L Brass et al. Cell. 2009.
Abstract
Influenza viruses exploit host cell machinery to replicate, resulting in epidemics of respiratory illness. In turn, the host expresses antiviral restriction factors to defend against infection. To find host cell modifiers of influenza A H1N1 viral infection, we used a functional genomic screen and identified over 120 influenza A virus-dependency factors with roles in endosomal acidification, vesicular trafficking, mitochondrial metabolism, and RNA splicing. We discovered that the interferon-inducible transmembrane proteins IFITM1, 2, and 3 restrict an early step in influenza A viral replication. The IFITM proteins confer basal resistance to influenza A virus but are also inducible by interferons type I and II and are critical for interferon's virustatic actions. Further characterization revealed that the IFITM proteins inhibit the early replication of flaviviruses, including dengue virus and West Nile virus. Collectively this work identifies a family of antiviral restriction factors that mediate cellular innate immunity to at least three major human pathogens.
Copyright 2009 Elsevier Inc. All rights reserved.
Figures
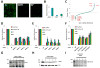
Fig. 1. The siRNA screen for influenza A virus infection modifying host factors
A) U2OS cells were transfected with the indicated siRNAs, then infected with influenza A virus (PR8) and immuno-stained 12 hours later for hemaggutinin (HA, green: HA). NP, siRNA targeting flu nucleoprotein, C, Nontargeting siRNA negative control. Magnification, 4×. B) Quantification of samples in A. Relative fold infection is normalized to non-targeting (C) control. Values represent the mean ± SD, N=4. C) The results of the screen are shown with the siRNA SMARTpools ranked in order of average Z-score, from lowest (decreased infection) to highest (increased infection). The position of known influenza A virus-host factors and several newly identified genes from the screen are indicated. D, E and F) U2OS cells were transfected with the indicated siRNAs for 72 h, and then infected with PR8. Twelve hours after infection the cells were analyzed by IF for the following viral proteins, HA (surface or entire cell), NP and M2. Relative fold infection is normalized to non-targeting (C) control. Values represent the mean ± SD, N =4. G, H and I) Western blots for cells in D, E and F. C, Non-targeting siRNA negative control. Ran levels are provided to demonstrate relative protein loading when cross-reacting bands were not present.
Fig. 2. Integrated model of influenza A virus host factors
Using the influenza A virus lifecycle as a guide (Lamb and Krug, 2001), the candidate proteins from the human and fly screens were placed at the position most likely to be relevant to the virus using a database of annotations from Gene Ontology, KEGG, Reactome and OMIM (see methods). Computational mapping and supporting evidences were reviewed and refined manually (Dataset S1C-F). The known molecular functions of the host factors were determined with the use of bioinformatics and multiple data sets (gray ovals). Host factors identified in the human siRNA screen (blue); the human orthologues of proteins identified in the fly-based screen (pink), factors which were found in both human and fly screens (green), and bridging proteins that were not detected, but none-the-less generate potentially insightful interactions (gray). Double borders signify the candidate is present in the Reactome influenza A virus infection pathway (Vastrik et al., 2007). Solid lines between genes indicate a protein interaction in human or other speicies. Dotted lines indicate inferred interaction from literature or annotation. Viral RNA (vRNA), viral complementary RNA (cRNA).
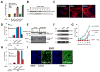
Fig. 3. IFITM3 silencing increases influenza A virus infection, and is required for the anti-viral actions of IFN
A) U2OS cells were transfected with the indicated siRNAs, then infected with PR8. Infection was assessed by IF for HA (surface or entire cell), NP or M2, 12 h after viral addition. Relative fold infection is normalized to non-targeting (C) control. Values represent the mean ± SD, N=3 throughout. B) U2OS cells transfected with the indicated siRNAs were assessed for IFITM3 levels by Western blotting. C) WI-38 human primary fibroblast cells stained for basal levels of IFITM3 expression (left panel), or after 24 h treatment with IFN-γ, or IFN-α (red: IFITM3, blue: nuclei), 63×. D) U2OS cells stably expressing either IFITM3 with a C-terminal HA-epitope tag (IFTIM3-HA6R) lacking the target site for siRNA IFITM3-6, or the vector alone, were transfected with the indicated siRNAs (x-axis). After 72 h the cells were incubated without (no virus) or with influenza A (PR8) for 12 h, then stained for HA expression. The anti-HA antibody used to detect infection does not recognize the HA epitope tag on IFITM3-HA6R (no virus, uninfected control). C, non-targeting siRNA negative control. E) U2OS cells stably expressing either IFTIM3-HA6R or vector alone were transfected with the indicated siRNAs and assessed 72 h after transfection by Western blot with the indicated antibodies. F) U2OS cells were untreated (-), or incubated with either IFN-γ, or IFN-α. After 24 h the levels of IFITM1, 2 or 3 were checked by Western blot. G) U2OS cells were transfected with the indicated siRNAs, then left untreated or incubated with IFN-γ 48 h later. After 24 h of IFN incubation, the cells were infected with increasing amounts of PR8. Twelve h after infection the cells were stained for HA expression. H) IFITM3 is required for the anti-viral effect of IFN-γ. U2OS cells stably expressing either IFTIM36R or vector, were transfected with the indicated siRNAs (x-axis) and treated with IFN-γ 48 h later. After 24 h, cells were incubated without or with influenza A (PR8) and 12 h after infection cells were checked for HA surface expression. I) IFITM3 loss enhances infection by the H1N1 strain, WS/33. U20S cells were transfected with the indicated siRNAs for 72 h, infected with WS/33 for 12 h, and stained for HA expression (green, blue: nuclei). Numbers indicate percent infection normalized to control /± SD, N=3.
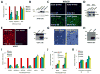
Fig. 4. The IFITM protein family restricts influenza A virus infection
A) A549 cells were transduced with retroviruses containing C9-tagged cDNAs for the indicated IFITM proteins, or empty viral vector alone. After 2 d cells were infected with one of the following viruses: PR8 [H1(PR)], influenza A virus A/Udorn/72 (H3N2) [H3(Udorn)], or Moloney murine leukemia virus (MLV). 24 h after infection, cells were checked for HA surface expression by flow cytometry. Values represent the mean ± SD, N=3. B) The expression of IFITM proteins in (A) was checked by Western using anti-C9 antibody. β-actin levels show protein loading. C) A549 or U2OS cells stably over-expressing IFITM3 protein or vector alone, were infected with influenza A H1N1 WSN/33. 12 h. later, cells were fixed and stained for surface HA expression. Values represent the mean /± SD, N=3. (green: HA, blue: nuclei. 4×). D) A549 and U2OS cells stably over-expressing IFITM3 were tested for expression by Western. E) Primary Chicken fibroblast cells (ChEFs) stably over-expressing IFITM3 protein or vector alone, were infected with influenza A H1N1 WSN/33. 12 h. later, cells were fixed and stained for surface HA expression. Values represent the mean /± SD, N=3. (red: HA, blue: nuclei. 4×). F) ChEF cells stably over-expressing IFITM3 were tested for expression by Western. G) MDCK cells stably over-expressing IFITM3 protein or vector alone, were infected with influenza A H1N1 WSN/33 at a multiplicity of infection (moi) of 0.005. 72 h. later, cells were washed with fresh media, and then imaged live to assess cytopathic effect. Bright field images shown are representative of 4 independent experiments (10×). H) MDCK cells stably over-expressing IFITM3 were tested for expression by Western. I) A549 cells were transduced with retroviruses containing the indicated IFITM proteins, or empty vector. 48 h later, the cells were incubated with MLV-EGFP virus pseudotyped with the indicated envelope proteins. HA proteins from various influenza A virus strain including H1 (PR): A/PR/8/34 (H1N1), H3 (Udorn): A/Udorn/72 (H3N2), H5(Thai): A/Thailand2(SP-33)/2004 (H5N1), H7(FPV): A/FPV/Rostock/34 (H7N1), VSV-G: VSV G protein, MLV: MLV env protein, or MACH: Machupo virus glycoprotein. Viral entry is expressed as mean EGFP fluorescence relative to vector control cells, as measured by flow cytometry. Values represent the mean ± SD, N=3. J) U2OS cells transfected with the indicated siRNAs for 72 h, were then incubated with MLV-GFP virus pseudotyped with the VSV-G or the HA protein of PR8, H1(PR). Entry, represented as percent green fluorescing cells relative to mock-transfected cells, was determined by IF microscopy two days post-infection. Values represent the mean ± SD, N=4. C, Non-targeting siRNA negative control. K) A549 cells were transduced with the indicated retroviruses. 48 h later, the cells were tested for surface expression of sialic acid (SA). Values represent the mean ± SD, N=3.
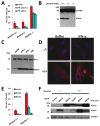
Fig. 5. Ifitm knockout cells are more susceptible to influenza A H1N1 virus infection, and are protected by the reinstatement of Ifitm2 or 3 expression
A) MEFs derived from the indicated IfitmDel mice, were left untreated (buffer), or treated with interferon-α or γ. After 24 h, the cells were incubated with influenza A virus H1N1 (PR8). Twelve h after infection, the cells were checked for HA surface expression. Values represent the mean ± SD, N=3. B) MEFs from A) were assessed by Western blot for the presence of Ifitm3 protein. GAPDH levels are provided to show protein loading. C) MEFs were left untreated (buffer), or incubated with either IFN-γ, IFN-α, or PR8 virus. After 24 h the levels of Ifitm3 were checked by Western blot. D) IfitmDel +/+ MEFs, or IfitmDel -/- MEFs were incubated in the absence (buffer) or presence of IFN-α for 24 h, prior to staining with α-Ifitm3 (red: Ifitm3, nuclei: blue). 63×. E) IfitmDel +/+ MEFs, or IfitmDel -/- MEFs stably expressing Ifitm2, 3 or the empty vector, were challenged with PR8 virus. 12 h later, the cells were fixed and imaged for HA expression. Values represent the mean ± SD, N=3. F) The indicated MEFS were assessed for Ifitm2 and 3 expression by Western blot. GAPDH demonstrates protein loading.
Fig. 6. The IFITM protein family restricts West Nile virus and dengue virus infections
A) Vero E6 cells were transduced with retroviruses expressing the indicated IFITM proteins, or the empty viral vector. Two days later, the cells were incubated with flaviviral viral like particles (VLPs), expressing EGFP, and coated in envelope proteins from WNV, yellow fever virus (YFV) or Omsk virus (OMSK), or with EGFP-expressing MLV viruses pseudotyped with the indicated viral envelope proteins. Viral infection is expressed as mean EGFP fluorescence relative to vector control cells, as measured by flow cytometry 48 h post-infection. Values represent the mean ± SD, N=3. B) A549 or U2OS cells stably expressing either IFITM3 protein or the vector alone (also shown in 4C, D), were infected with infectious WNV (strain 2741). 24 h. later, the cells were fixed and stained for viral E protein expression by IF. Values represent the mean ± SD, N=3 C) Images of A549 cells in
B
(red: WNV E protein, blue: nuclei), 4× magnification. D) HeLa cells were transfected with the indicated siRNAs for 72 h, then infected with WNV. 24 h later, the cells were fixed and stained for viral E protein. Values represent the mean ± SD, N=3. C, Non-targeting siRNA negative control. E) Images of HeLa cells in D (red: WNV E protein, blue: nuclei), 4× magnification. F) HeLa cells were transfected with the indicated siRNAs for 72 h, then infected with dengue virus (New Guinea C strain). 30 h. post-infection, the cells were fixed and stained for viral E protein expression by IF. Values represent the mean ± SD, N=3. C, Non-targeting siRNA negative control. G) Images of HeLa cells in F, 4×.
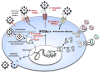
Fig. 7. IFITM proteins act as anti-viral restriction factors
A schematic model of the influenza A virus life cycle, the induction of IFITM proteins and their role in blocking Influenza A virus infection. IFITM1, 2 and 3 are represented by the three multi-transmembrane proteins. Red legends indicate possible mechanisms of restriction, IFITM proteins may; A) sequester the incoming viruses at or near the surface, B) block viral receptors from interacting with host receptors (shown in orange), C) prevent endocytosis or viral membrane fusion D) act as receptors, and after binding the virus, signal to effectors.
Similar articles
- IFITM Proteins That Restrict the Early Stages of Respiratory Virus Infection Do Not Influence Late-Stage Replication.
Meischel T, Fritzlar S, Villalon-Letelier F, Tessema MB, Brooks AG, Reading PC, Londrigan SL. Meischel T, et al. J Virol. 2021 Sep 27;95(20):e0083721. doi: 10.1128/JVI.00837-21. Epub 2021 Jul 28. J Virol. 2021. PMID: 34319159 Free PMC article. - IFITM proteins restrict antibody-dependent enhancement of dengue virus infection.
Chan YK, Huang IC, Farzan M. Chan YK, et al. PLoS One. 2012;7(3):e34508. doi: 10.1371/journal.pone.0034508. Epub 2012 Mar 30. PLoS One. 2012. PMID: 22479637 Free PMC article. - Long Noncoding RNA IFITM4P Regulates Host Antiviral Responses by Acting as a Competing Endogenous RNA.
Xiao M, Chen Y, Wang S, Liu S, Rai KR, Chen B, Li F, Li Y, Maarouf M, Chen JL. Xiao M, et al. J Virol. 2021 Oct 13;95(21):e0027721. doi: 10.1128/JVI.00277-21. Epub 2021 Jul 21. J Virol. 2021. PMID: 34287042 Free PMC article. - [Research Progress on Antiviral Activity of Interferon-induced Transmembrane Proteins].
Chen Y, Zhu W, Shu Y. Chen Y, et al. Bing Du Xue Bao. 2016 Mar;32(2):222-8. Bing Du Xue Bao. 2016. PMID: 27396168 Review. Chinese. - Modulation of Innate Immune Responses by the Influenza A NS1 and PA-X Proteins.
Nogales A, Martinez-Sobrido L, Topham DJ, DeDiego ML. Nogales A, et al. Viruses. 2018 Dec 12;10(12):708. doi: 10.3390/v10120708. Viruses. 2018. PMID: 30545063 Free PMC article. Review.
Cited by
- A Functional Role of Fibroblast Growth Factor Receptor 1 (FGFR1) in the Suppression of Influenza A Virus Replication.
Liu X, Lai C, Wang K, Xing L, Yang P, Duan Q, Wang X. Liu X, et al. PLoS One. 2015 Apr 24;10(4):e0124651. doi: 10.1371/journal.pone.0124651. eCollection 2015. PLoS One. 2015. PMID: 25909503 Free PMC article. - Chicken interferon-induced transmembrane protein 1 promotes replication of coronavirus infectious bronchitis virus in a cell-specific manner.
Li H, Ni R, Wang K, Tian Y, Gong H, Yan W, Tang Y, Lei C, Wang H, Yang X. Li H, et al. Vet Microbiol. 2022 Dec;275:109597. doi: 10.1016/j.vetmic.2022.109597. Epub 2022 Oct 28. Vet Microbiol. 2022. PMID: 36368134 Free PMC article. - The Interferon-Stimulated Gene IFITM3 Restricts Infection and Pathogenesis of Arthritogenic and Encephalitic Alphaviruses.
Poddar S, Hyde JL, Gorman MJ, Farzan M, Diamond MS. Poddar S, et al. J Virol. 2016 Sep 12;90(19):8780-94. doi: 10.1128/JVI.00655-16. Print 2016 Oct 1. J Virol. 2016. PMID: 27440901 Free PMC article. - Widespread but tissue-specific patterns of interferon-induced transmembrane protein 3 (IFITM3, FRAGILIS, MIL-1) in the mouse gastrula.
Mikedis MM, Downs KM. Mikedis MM, et al. Gene Expr Patterns. 2013 Oct;13(7):225-39. doi: 10.1016/j.gep.2013.04.003. Epub 2013 Apr 29. Gene Expr Patterns. 2013. PMID: 23639725 Free PMC article. - Replication-Competent Influenza A Viruses Expressing Reporter Genes.
Breen M, Nogales A, Baker SF, Martínez-Sobrido L. Breen M, et al. Viruses. 2016 Jun 23;8(7):179. doi: 10.3390/v8070179. Viruses. 2016. PMID: 27347991 Free PMC article. Review.
References
- Alber D, Staeheli P. Partial inhibition of vesicular stomatitis virus by the interferon-induced human 9-27 protein. J Interferon Cytokine Res. 1996;16:375–380. - PubMed
- Boulo S, Akarsu H, Ruigrok RW, Baudin F. Nuclear traffic of influenza virus proteins and ribonucleoprotein complexes. Virus Res. 2007;124:12–21. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AI062773/AI/NIAID NIH HHS/United States
- DK043351/DK/NIDDK NIH HHS/United States
- P30 DK040561/DK/NIDDK NIH HHS/United States
- R01 DK060049/DK/NIDDK NIH HHS/United States
- U01 AI070343/AI/NIAID NIH HHS/United States
- P30 DK043351/DK/NIDDK NIH HHS/United States
- U54 AI057159/AI/NIAID NIH HHS/United States
- DK060049/DK/NIDDK NIH HHS/United States
- R01 AI062773/AI/NIAID NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- CRUK_/Cancer Research UK/United Kingdom
- AI070343/AI/NIAID NIH HHS/United States
- AI 50031/AI/NIAID NIH HHS/United States
- N01 AI050031/AI/NIAID NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases