Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells - PubMed (original) (raw)
Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells
Xiaohua Shen et al. Cell. 2009.
Abstract
Trimethylation on histone H3 lysine 27 (H3K27me3) by Polycomb repressive complex 2 (PRC2) regulates the balance between self-renewal and differentiation of embryonic stem cells (ESCs). The mechanisms controlling the activity and recruitment of PRC2 are largely unknown. Here we demonstrate that the founding member of the Jumonji family, JMJ (JUMONJI or JARID2), is associated with PRC2, colocalizes with PRC2 and H3K27me3 on chromatin, and modulates PRC2 function. In vitro JMJ inhibits PRC2 methyltransferase activity, consistent with increased H3K27me3 marks at PRC2 targets in Jmj(-/-) ESCs. Paradoxically, JMJ is required for efficient binding of PRC2, indicating that the interplay of PRC2 and JMJ fine-tunes deposition of the H3K27me3 mark. During differentiation, activation of genes marked by H3K27me3 and lineage commitments are delayed in Jmj(-/-) ESCs. Our results demonstrate that dynamic regulation of Polycomb complex activity orchestrated by JMJ balances self-renewal and differentiation, highlighting the involvement of chromatin dynamics in cell-fate transitions.
Copyright 2009 Elsevier Inc. All rights reserved.
Figures
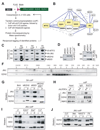
Figure 1. JMJ interacts with the PRC2 complex
(A) The scheme to identify PRC2 interacting proteins. (B) A PRC2 interactome. Proteins highlighted in yellow are tagged baits for affinity purification. Solid and dotted lines indicate interactions identified by multiple or few replicates, respectively. The interaction between BCLAF1 and THRAP3 was reported previously (Bracken et al., 2008). (C) – (E) Coimmunoprecipitation (coIP) analysis in 293T cells. V5-tagged MTF2, HA-tagged JMJ, bfEED and bfEZH2 were coexpressed (C). V5-MTF2 (D) or HA-JMJ (E) was coexpressed with bfEED, bfEZH1, or bfEZH2 as indicated. (F) Gel filtration analysis of nuclear extracts from CJ7 ESCs. Migration of molecular markers is indicated above the panels, and the antibodies for Western blotting are shown on the left. (G) Streptavidin (SA) mediated coIP in ESCs expressing birA alone, bfEED, bfEZH2 or mutant EZH2 with SET domain deleted (bfEZH2dSET). bfEZH2dSET is indicated by *. (H) Anti-SUZ12 coIP in Jmfl/fl and _Jmj_−/− ESCs. (I) – (J) Anti-JMJ antibody-mediated coIP in ESCs and non-pluripotent cells. In (C) – (J), “I” indicates inputs and “P” indicates coIP fractions. About 5% of nuclear extracts used for coIP were loaded as the inputs.
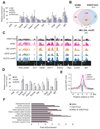
Figure 2. JMJ colocalizes with the PRC2 and H3K27me3 mark on chromatin
(A) ChIP-qPCR analysis in ESCs. The Actb promoter and an intergenic region (Cs) serve as negative controls. Error bars represent standard deviations of relative enrichments to inputs. (B) Venn diagram of genome-wide co-localization of JMJ, bfJMJ and the H3K27me3 mark. (C) Representative view of ChIP-chip peaks at various genomic loci in the Affymetrix Integrated Genome Browser. Arrows indicate transcription start and direction. Unless indicated by “mmP” (Affymetrix mouse promoter tiling array), chip hybridization was performed on mouse whole genome tiling array sets. (D) BioChIP-qPCR analysis in ESCs expressing birA control only, bfJMJ or bfEZH2. Error bars represent standard deviations of relative enrichments to inputs. (E) ChIP-chip signals around transcription start sites (TSS). Average enrichment ratios within +/− 10kb regions relative to TSS were calculated based on MAT scores. (F) Gene ontology (GO) analysis of H3K27me3 target genes (p < 1e-5).
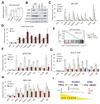
Figure 3. Histone methyltransferase assays show intrinsic KMT activity within the JMJ-containing complex
(A) Addition of increasing amounts of the JMJ-containing complex captured by streptavidin (SA)-mediated coIP from ESCs expressing bfJMJ exhibits increasing KMT activity on chicken core histones. BirA and bfEZH2 serve as the negative and positive control, respectively. (B) Proteins captured by anti-JMJ and anti-SUZ12 antibodies show KMT activity toward nucleosomes from Hela cells. Nucleosomes were pretreated at 50°C for 10 minutes to inactivate copurified, endogenous KMTs. (C) KMT activity of bfJMJ and bfEZH2 complexes on recombinant histone H3 substrates. The bottom panel shows Western blotting of EZH2 to ensure equal addition of coIP’ed protein complexes. (D) KMT activity of endogenous JMJ protein complex captured by an anti-JMJ antibody on recombinant histone H3 substrates. The lower panels in (A), (B) and (D) and the middle panel in (C) are protein gels stained by Colloidal blue to show equal addition of substrates.
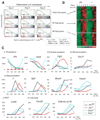
Figure 4. JMJ inhibits methyltransferase activity of PRC2
(A) Reconstitution of JMJ with PRC2 in vitro. Except for the first lane in which JMJ-expressing baculovirus is omitted, FLAG-tagged JMJ was coexpressed with FLAG-tagged EZH2, HA-tagged EED, SUZ12, RBBP4 and AEBP2 in Sf21 cells. Captured proteins were separated on the gel and stained by Colloidal blue. While equal amounts of JMJ and EZH2 were captured by anti-FLAG antibody, JMJ captured with HA-EED is one-fourth the amount of EZH2, indicating JMJ as a substoichiometric component of PRC2. (B) JMJ inhibits intrinsic methyltransferase activity associated with crude nucleosomes purified from Hela cells. The top plot shows the quantification of the autoradiography (in the middle). The lower panel shows equal loading of nucleosomes (5 µg). About 0.5 – 2 µg of JMJ and 4 – 10 µg of bovine serum albumin (BSA) were added as indicated. (C) JMJ inhibits di- and tri-methyltransferase activities of PRC2 on recombinant histones (1 µg per reaction). About 2 – 10 µg of BSA (indicated by #) was added as indicated. (D) The JmjC and C5HC2 domains of JMJ are dispensable for the inhibitory function of JMJ on PRC2. The protein band corresponding to the JMJ mutant (dJc) is indicated by *. (E) – (F) JMJ inhibits all methylation activities of PRC2 on MLA octamers (E) and nucleosomes (F). About 2 µg of BSA was added. In (C) – (F), 5 µg of PRC2 and 0.5 µg – 2 µg of JMJ or dJc were added as indicated. The upper panels show autoradiography. The middle and bottom panels are protein gels stained by Colloidal blue to show additions of substrates and enzymes, respectively.
Figure 5. Characterization of _Jmj_−/− ESCs
(A) Reverse transcription (RT) and qPCR analysis of Jmj transcripts in wild-type and mutant ESCs. Error bars are standard deviations of relative expression to GADPH. (B) Western blotting analysis of wild-type and mutant ESCs. (C) ChIP-qPCR analysis of JMJ in wild-type (WT) and mutant ESCs. (D) ChIP-qPCR analysis of the H3K27me3 mark in _Jmj_−/− and wild-type ESCs. * indicates p < 0.05 by a Student's paired t-Test with a two-tailed distribution. (E) GSEA profile of the set of H3K27me3 target genes. Genes are ranked into an ordered list based on the correlation between their expression levels in _Jmj_−/− and Jmjfl/fl ESCs. Genes which are upregulated in _Jmj_−/− ESCs are ranked at the top of the list (left, red), while downregulated genes are ranked toward the bottom (right, blue). The middle portion of the plot shows where the members of the gene set (indicated by vertical lines) locate in the ranked list shown in the bottom portion. The green curve in the top portion shows the running enrichment score (ES) for the gene set as the analysis walks down the ranked list. The distribution of H3K27me3 target genes appear toward the bottom of the rank list with a normalized enrichment score (NES) of −2.9. Statistic significance is indicated by nominal p value, familywise-error rate (FWER) and false discovery rate (FDR). (F) – (H) ChIP-qPCR analysis of EZH2 (F), SUZ12 (G) and EED (H) at PRC2 targets in wild-type and mutant ESCs. In (C), (D) and (F)–(H), error bars are standard deviations of relative enrichments based on at least three biological repeats. The Actb promoter and an intergenic region (Cs) serve as negative controls. (I) A model shows dynamic regulation of H3K27me3. PRC2, the H3K27 methyltransferase complex (KMT), and UTX and JMJD3, the H3K27me3 demethylases (KDM), represent on-off controls on H3K27me3. While PRC2 catalyzes the formation of H3K27me3, UTX and JMJD3 demethylate H3K27me3. In contrast to KMTs and KDMs, JMJ, as a modifier of PRC2, fine-tunes the levels of H3K27me3 by regulating the activity and recruitment of PRC2.
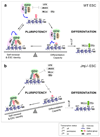
Figure 6. Jmj is required for ESC differentiation
(A) GSEA profiles of the sets of H3K27me3 target genes, NP-high (enriched in neuronal progenitors) and ME-high genes (enriched in mesoendodermal lineage). (B) Heatmaps show expression of genes in three gene sets indicated in (A) in Jmjfl/fl and _Jmj_−/− cells during ESC differentiation. (C) Time-course analysis of marker genes during differentiation by RT-qPCR. Error bars are standard deviations of relative expression shown in the y-axis.
Figure 7. A model for the interplay between JMJ and PRC2 in regulating ESC pluripotency
(A) In wild-type ESC, the inhibitory role of JMJ on PRC2 activity disfavors formation of H3K27me3 and may facilitate attacks by opposing enzyme or factors, such as UTX, JMJD3 and MLLs, resulting in transient H3K27me3 demethylation and a permissive chromatin structure for transcription. Paradoxically, the positive role of JMJ in promoting PRC2 binding may redirect PRC2 to chromatin to establish the H3K27me3 mark. Dynamic control of the H3K27me3 mark defines the fine balance between the maintenance of the ESC state and differentiation capacity, and is required for rapid relief of repression and subsequent gene activation upon differentiation signals. (B) In _Jmj_−/− ESCs, H3K27me3 may be more rigidly positioned at target loci. The balance tips towards maintenance of the stem cell phenotype. Genes marked and repressed by H3K27me3 become refractory to activation on differentiation signals.
Similar articles
- Polycomb-like 3 promotes polycomb repressive complex 2 binding to CpG islands and embryonic stem cell self-renewal.
Hunkapiller J, Shen Y, Diaz A, Cagney G, McCleary D, Ramalho-Santos M, Krogan N, Ren B, Song JS, Reiter JF. Hunkapiller J, et al. PLoS Genet. 2012;8(3):e1002576. doi: 10.1371/journal.pgen.1002576. Epub 2012 Mar 15. PLoS Genet. 2012. PMID: 22438827 Free PMC article. - Jarid2 and PRC2, partners in regulating gene expression.
Li G, Margueron R, Ku M, Chambon P, Bernstein BE, Reinberg D. Li G, et al. Genes Dev. 2010 Feb 15;24(4):368-80. doi: 10.1101/gad.1886410. Epub 2010 Feb 1. Genes Dev. 2010. PMID: 20123894 Free PMC article. - Jarid2 Methylation via the PRC2 Complex Regulates H3K27me3 Deposition during Cell Differentiation.
Sanulli S, Justin N, Teissandier A, Ancelin K, Portoso M, Caron M, Michaud A, Lombard B, da Rocha ST, Offer J, Loew D, Servant N, Wassef M, Burlina F, Gamblin SJ, Heard E, Margueron R. Sanulli S, et al. Mol Cell. 2015 Mar 5;57(5):769-783. doi: 10.1016/j.molcel.2014.12.020. Epub 2015 Jan 22. Mol Cell. 2015. PMID: 25620564 Free PMC article. - Polycomb repressive complex 2 in embryonic stem cells: an overview.
Jones A, Wang H. Jones A, et al. Protein Cell. 2010 Dec;1(12):1056-62. doi: 10.1007/s13238-010-0142-7. Epub 2011 Jan 8. Protein Cell. 2010. PMID: 21213100 Free PMC article. Review. - The Polycomb complex PRC2 and its mark in life.
Margueron R, Reinberg D. Margueron R, et al. Nature. 2011 Jan 20;469(7330):343-9. doi: 10.1038/nature09784. Nature. 2011. PMID: 21248841 Free PMC article. Review.
Cited by
- A complex Polycomb issue: the two faces of EZH2 in cancer.
Hock H. Hock H. Genes Dev. 2012 Apr 15;26(8):751-5. doi: 10.1101/gad.191163.112. Genes Dev. 2012. PMID: 22508723 Free PMC article. - REST-mediated recruitment of polycomb repressor complexes in mammalian cells.
Dietrich N, Lerdrup M, Landt E, Agrawal-Singh S, Bak M, Tommerup N, Rappsilber J, Södersten E, Hansen K. Dietrich N, et al. PLoS Genet. 2012;8(3):e1002494. doi: 10.1371/journal.pgen.1002494. Epub 2012 Mar 1. PLoS Genet. 2012. PMID: 22396653 Free PMC article. - EED and KDM6B coordinate the first mammalian cell lineage commitment to ensure embryo implantation.
Saha B, Home P, Ray S, Larson M, Paul A, Rajendran G, Behr B, Paul S. Saha B, et al. Mol Cell Biol. 2013 Jul;33(14):2691-705. doi: 10.1128/MCB.00069-13. Epub 2013 May 13. Mol Cell Biol. 2013. PMID: 23671187 Free PMC article. - Developmental control of polycomb subunit composition by GATA factors mediates a switch to non-canonical functions.
Xu J, Shao Z, Li D, Xie H, Kim W, Huang J, Taylor JE, Pinello L, Glass K, Jaffe JD, Yuan GC, Orkin SH. Xu J, et al. Mol Cell. 2015 Jan 22;57(2):304-316. doi: 10.1016/j.molcel.2014.12.009. Epub 2015 Jan 8. Mol Cell. 2015. PMID: 25578878 Free PMC article. - Integrative genomic analysis of early neurogenesis reveals a temporal genetic program for differentiation and specification of preplate and Cajal-Retzius neurons.
Li J, Sun L, Peng XL, Yu XM, Qi SJ, Lu ZJ, Han JJ, Shen Q. Li J, et al. PLoS Genet. 2021 Mar 24;17(3):e1009355. doi: 10.1371/journal.pgen.1009355. eCollection 2021 Mar. PLoS Genet. 2021. PMID: 33760820 Free PMC article.
References
- Ambrosi DJ, Tanasijevic B, Kaur A, Obergfell C, O'Neill RJ, Krueger W, Rasmussen TP. Genome-wide reprogramming in hybrids of somatic cells and embryonic stem cells. Stem cells (Dayton, Ohio) 2007;25:1104–1113. - PubMed
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. - PubMed
- Chambers I, Silva J, Colby D, Nichols J, Nijmeijer B, Robertson M, Vrana J, Jones K, Grotewold L, Smith A. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases