LRRTM2 interacts with Neurexin1 and regulates excitatory synapse formation - PubMed (original) (raw)
LRRTM2 interacts with Neurexin1 and regulates excitatory synapse formation
Joris de Wit et al. Neuron. 2009.
Abstract
We identify the leucine-rich repeat transmembrane protein LRRTM2 as a key regulator of excitatory synapse development and function. LRRTM2 localizes to excitatory synapses in transfected hippocampal neurons, and shRNA-mediated knockdown of LRRTM2 leads to a decrease in excitatory synapses without affecting inhibitory synapses. LRRTM2 interacts with PSD-95 and regulates surface expression of AMPA receptors, and lentivirus-mediated knockdown of LRRTM2 in vivo decreases the strength of evoked excitatory synaptic currents. Structure-function studies indicate that LRRTM2 induces presynaptic differentiation via the extracellular LRR domain. We identify Neurexin1 as a receptor for LRRTM2 based on affinity chromatography. LRRTM2 binds to both Neurexin 1alpha and Neurexin 1beta, and shRNA-mediated knockdown of Neurexin1 abrogates LRRTM2-induced presynaptic differentiation. These observations indicate that an LRRTM2-Neurexin1 interaction plays a critical role in regulating excitatory synapse development.
2009 Elsevier Inc. All rights reserved.
Figures
Figure 1
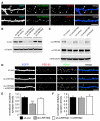
Knockdown of LRRTM2 Decreases Excitatory Synapse Density in Hippocampal Neurons (A) Hippocampal neurons were cotransfected with myc-LRRTM2 and GFP at DIV 10 and immunostained for postsynaptic markers and GFP at DIV 17. Top panels, myc-LRRTM2 colocalizes with the excitatory postsynaptic marker PSD-95 in dendritic spines (arrowheads). Bottom panels, myc-LRRTM2 does not colocalize with the inhibitory postsynaptic marker gephyrin. GFP fluorescence shown in blue for better visualization. (B) 293T cells were cotransfected with mouse myc-LRRTM2 and the empty pSUPER vector (sh-vector) or pSUPER containing a shRNA against mouse and rat LRRTM2 (sh-LRRTM2) for 48 hr. The LRRTM2 shRNA reduces expression of mouse LRRTM2 by 90%, but human myc-LRRTM2 is not affected. Samples were probed with a β-actin antibody to verify equal loading. (C) Knockdown of endogenous LRRTM2 in hippocampal neurons. Neurons were infected with control or sh-LRRTM2 containing lentiviral vectors (LV) at DIV 6 and analyzed at DIV 12. LRRTM2 protein levels are strongly reduced in LV-shLRRTM2-infected neurons compared to control infected or uninfected neurons. PSD-95 and βIII tubulin levels in the same samples are unchanged. (D) Hippocampal neurons were electroporated with the sh-vector (sh-vec), sh-LRRTM2, and sh-LRRTM2 with human myc-LRRTM2 (sh-LRRTM2 + hLRRTM2) and immunostained at DIV 14 for GFP, PSD-95, and VGlut1. (E) Quantification of the density of VGlut1/PSD-95-positive puncta per length of dendrite normalized to sh-vec control neurons. (***p < 0.001). (F) LRRTM2 knockdown does not affect inhibitory synapse density identified by VGAT/gephyrin colocalization. Scale bar in (A) and (D), 10 μm.
Figure 2
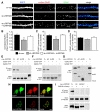
LRRTM2 Regulates GluR1 Surface Expression in Hippocampal Neurons and Interacts with Postsynaptic Proteins (A) Hippocampal neurons were electroporated with the sh-vector (sh-vec), sh-LRRTM2, and sh-LRRTM2 with human myc-LRRTM2 (sh-LRRTM2 + hLRRTM2), surface labeled for GluR1, and immunostained at DIV 15 for GFP and VGlut1. (B) Quantification of the density of synaptic surface GluR1 puncta (VGlut1/GluR1 colocalization) per length of dendrite normalized to sh-vec control neurons (**p < 0.001). (C) Quantification of the GluR1 surface puncta per length of dendrite (**p < 0.01). (D) Quantification of the VGlut1 puncta per 10 μm dendrite sh-vec 0.68 ± 0.03 (mean ± SEM); sh-LRRTM2 0.59 ± 0.02; sh-LRRTM2 + hLRRTM2 0.70 ± 0.04. Bar graphs show mean ± SEM (difference not significant by ANOVA; p = 0.02 by Student’s t test for control versus sh-LRRTM2). (E–G) 293T cells were cotransfected with myc-LRRTM2 mutant constructs and GluR1-GFP (E) or GluR2-GFP (F). Cell lysates were immunoprecipitated with myc antibodies and were analyzed by western blot using anti-GFP antibodies. EphB2-YFP was used as a negative control. A representative blot showing input samples probed with myc antibody is shown in (G). High and low molecular weight bands for LRRTM2 on western blot are probably due to glycosylation of LRRTM2. (H) Coexpression of PSD-95-mCherry with myc-LRRTM2 in 293T cells induces translocation to the cell membrane of PSD-95, which is diffusely distributed throughout the cytosol in GFP-expressing control cells. (I) Immunoprecipitation of myc-LRRTM2 deletion mutants lacking the cytoplasmic domain (LRRTM2 ΔC) or the C-terminal ECEV motif (LRRTM2 ΔECEV) coexpressed with PSD-95-mCherry in 293T cells. LRRTM2 interacts with PSD-95-mCherry via the ECEV motif. The high molecular weight band is shown for LRRTM2. Molecular weight markers in kDa indicated on the right. Scale bar in (A) and (H), 10 μm.
Figure 3
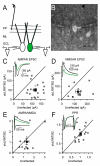
LRRTM2 Is Required for Excitatory Synapse Function In Vivo (A) Schematic of experimental configuration. Rats were stereotaxically injected with lentivirus at P5, and acute slices were cut between P13 and P16. Whole-cell recordings were made from nearby infected and uninfected granule cells, identified by DIC and GFP epifluorescence, in the granule cell layer (GCL). Perforant path (PP) inputs were stimulated with an electrode in the outer half of the molecular layer (ML). (B) Overlaid GFP epifluorescence and DIC images of a simultaneous recording. (C) LRRTM2 shRNA causes a large reduction in AMPAR-mediated EPSCs (average AMPAR EPSC uninfected 119.5 ± 7.7 pA [mean ± SEM]: sh-LRRTM2 50.7 ± 8.5: n = 17 pairs, p < 0.0001). For all scatter plots, open symbols represent means from individual experiments and the filled symbol represents the group mean ± SEM. Inset: example average evoked PP AMPAR EPSCs recorded simultaneously from shLRRTM2 infected (green) and uninfected (black) GCs at a holding potential of −60 mV. (D) LRRTM2 shRNA causes a large reduction in NMDAR-mediated EPSCs (average NMDAR EPSC uninfected 129.0 ± 18.7 pA [mean ± SEM]: sh-LRRTM2 59.7 ± 13.5: n = 11 pairs, p = 0.005). Inset: example average evoked PP compound AMPAR- and NMDAR-mediated EPSCs recorded at a holding potential of +40 mV. Arrows indicate the time point 50 ms after the stimulus at which the NMDAR-mediated amplitude was measured. (E) LRRTM2 shRNA causes a slight reduction in AMPA/NMDA ratio (average PP AMPA/NMDA ratio uninfected 1.32 ± 0.21 [mean ± SEM]: sh-LRRTM2 1.06 ± 0.30: n = 11 pairs, p = 0.04). Inset: example average overlaid AMPAR and NMDAR EPSCs with traces scaled to the peak of the AMPAR EPSC. (F) LRRTM2 shRNA does not affect the paired-pulse ratio (PPR) of PP inputs onto GCs (averaged PP paired-pulse ratios uninfected 1.05 ± 0.06 [mean ± SEM]: sh-LRRTM2 1.01 ± 0.08: n = 17 pairs, p = 0.54). Inset: example average PP EPSCs from a simultaneous recording. Stimuli were delivered at 20 Hz. Traces are scaled to the peak of the first EPSC.
Figure 4
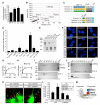
Neurexins Are Functional LRRTM2 Receptors (A) LRRTM2 FL and LRRTM2 ΔC expressed in hippocampal neurons significantly increase excitatory synapse density (normalized excitatory synapse density) (***p < 0.001; ΔLRR not significantly different from controls by ANOVA, but p = 0.01 for pairwise comparison with control by Student’s t test). See Figure S6E for accompanying fluorescence images. (B) Mass spec interaction screen identifies Neurexins as potential LRRTM2 receptors. The scatter plot is a graphical representation of a representative interaction screen. Each dot represents a protein that was identified by tandem mass spectrometry. The blue dot is LRRTM2 (bait protein), while the red dots are Neurexin family members. Gray dots represent other proteins that were also identified. The x axis represents the number of peptides and the y axis is the number of spectra in which the identified protein was found. (C) Schematic representation of Neurexin constructs. Neurexins (Nrxn) 1α, 2α, 3α, and 1β refer to the wild-type proteins. Nrxn 2α1β and Nrxn 3α1β are chimeric proteins of Nrxn1β with the LNS6 domain replaced with that of 2α and 3α, respectively. LNS, laminin, neurexin, sex-hormone-binding protein; CH, highly glycosylated region; βN, β specific leader; TM, transmembrane domain. All constructs have a C-terminal CFP tag. (D) Relative binding of LRRTM2-ecto-Fc to 293T cells expressing either Neurexins or control constructs. LRRTM2-Fc binds preferentially to 293T cells expressing Nrxn 1α, 1β, and 2α1β (*p < 0.05, **p < 0.005). All samples were normalized to an Fc-only control. (E) Relative expression of Neurexin-CFP constructs expressed in 293T cells, detected on western blot with anti-GFP antibody. (F) Confocal images of LRRTM2-ecto-Fc binding (red) to 293T cells (blue) expressing either Neurexin-CFP or control constructs. The CFP tag on neurexin constructs is pseudocolored blue to facilitate visualization of binding. Scale bar, 20 μm. (G) Binding of LRRTM2-ecto-Fc to Neurexin 1α-expressing 293T cells as a function of LRRTM2 concentration. (H) Binding of LRRTM2-ecto-Fc to Neurexin 1β-expressing 293T cells as a function of LRRTM2 concentration. (I and J) Direct binding of purified Neurexin 1α and 1β to immobilized LRRTM2-ecto-Fc. SDS-PAGE stained with Coomassie blue was used to separate the purified recombinant LRRTM2-ecto-Fc and either Neurexin 1α or 1β. Molecular weight standards, in kDa, are on the left. Proteins bands are identified on the right. FT, flow through of either Neurexin 1α (I) or Neurexin 1β (J). W1 through W4, washes; E1 through E3, elutions. Lack of binding to Fc control protein (E1–E3) is shown at bottom. Bait protein is LRRTM2-ecto-Fc. (K) Hippocampal neurons transfected with GFP (left) or GFP + shNRXN1 (right), cocultured with 293T cells expressing GFP + myc-LRRTM2. Cocultures were immunostained for GFP to identify axon/293T overlap and synapsin to identify presynaptic terminals. Arrowheads mark the trajectory of transfected axons. Note presence of synapsin puncta in control transfected axons (yellow) but not in shNrxn1-expressing axons. Scale bar, 10 μm. (L) Quantification of the experiment described in (K), showing that expression of shNrxn1 blocks the ability of LRRTM2 to induce presynaptic differentiation (*p < 0.05). Inset shows knockdown on Neurexin 1β, but not β-actin, in 293T cell lysates cotransfected with shNrxn1. (M) Model of regulation of excitatory synapse formation by LRRTM2-Neurexin1 interaction.
Similar articles
- LRRTM2 functions as a neurexin ligand in promoting excitatory synapse formation.
Ko J, Fuccillo MV, Malenka RC, Südhof TC. Ko J, et al. Neuron. 2009 Dec 24;64(6):791-8. doi: 10.1016/j.neuron.2009.12.012. Neuron. 2009. PMID: 20064387 Free PMC article. - Deletion of LRRTM1 and LRRTM2 in adult mice impairs basal AMPA receptor transmission and LTP in hippocampal CA1 pyramidal neurons.
Bhouri M, Morishita W, Temkin P, Goswami D, Kawabe H, Brose N, Südhof TC, Craig AM, Siddiqui TJ, Malenka R. Bhouri M, et al. Proc Natl Acad Sci U S A. 2018 Jun 5;115(23):E5382-E5389. doi: 10.1073/pnas.1803280115. Epub 2018 May 21. Proc Natl Acad Sci U S A. 2018. PMID: 29784826 Free PMC article. - Leucine-rich repeat transmembrane proteins are essential for maintenance of long-term potentiation.
Soler-Llavina GJ, Arstikaitis P, Morishita W, Ahmad M, Südhof TC, Malenka RC. Soler-Llavina GJ, et al. Neuron. 2013 Aug 7;79(3):439-46. doi: 10.1016/j.neuron.2013.06.007. Neuron. 2013. PMID: 23931994 Free PMC article. - SynDIG1 regulation of excitatory synapse maturation.
Díaz E. Díaz E. J Physiol. 2012 Jan 1;590(1):33-8. doi: 10.1113/jphysiol.2011.213884. Epub 2011 Aug 30. J Physiol. 2012. PMID: 21878521 Free PMC article. Review. - Promiscuous interactions between AMPA-Rs and MAGUKs.
Fitzjohn SM, Doherty AJ, Collingridge GL. Fitzjohn SM, et al. Neuron. 2006 Oct 19;52(2):222-4. doi: 10.1016/j.neuron.2006.10.002. Neuron. 2006. PMID: 17046684 Review.
Cited by
- Presynaptic neurexin-3 alternative splicing trans-synaptically controls postsynaptic AMPA receptor trafficking.
Aoto J, Martinelli DC, Malenka RC, Tabuchi K, Südhof TC. Aoto J, et al. Cell. 2013 Jul 3;154(1):75-88. doi: 10.1016/j.cell.2013.05.060. Cell. 2013. PMID: 23827676 Free PMC article. - Synaptic Organizers in Alzheimer's Disease: A Classification Based on Amyloid-β Sensitivity.
Lee AK, Khaled H, Chofflet N, Takahashi H. Lee AK, et al. Front Cell Neurosci. 2020 Sep 2;14:281. doi: 10.3389/fncel.2020.00281. eCollection 2020. Front Cell Neurosci. 2020. PMID: 32982693 Free PMC article. Review. - Distinct Alterations in Dendritic Spine Morphology in the Absence of β-Neurexins.
Mohrmann L, Seebach J, Missler M, Rohlmann A. Mohrmann L, et al. Int J Mol Sci. 2024 Jan 20;25(2):1285. doi: 10.3390/ijms25021285. Int J Mol Sci. 2024. PMID: 38279285 Free PMC article. - Distinct roles of neuroligin-1 and SynCAM1 in synapse formation and function in primary hippocampal neuronal cultures.
Burton SD, Johnson JW, Zeringue HC, Meriney SD. Burton SD, et al. Neuroscience. 2012 Jul 26;215:1-16. doi: 10.1016/j.neuroscience.2012.04.047. Epub 2012 Apr 25. Neuroscience. 2012. PMID: 22542674 Free PMC article. - Differences in AMPA and kainate receptor interactomes facilitate identification of AMPA receptor auxiliary subunit GSG1L.
Shanks NF, Savas JN, Maruo T, Cais O, Hirao A, Oe S, Ghosh A, Noda Y, Greger IH, Yates JR 3rd, Nakagawa T. Shanks NF, et al. Cell Rep. 2012 Jun 28;1(6):590-8. doi: 10.1016/j.celrep.2012.05.004. Epub 2012 May 23. Cell Rep. 2012. PMID: 22813734 Free PMC article.
References
- Biederer T, Scheiffele P. Mixed-culture assays for analyzing neuronal synapse formation. Nat. Protoc. 2007;2:670–676. PubMed. - PubMed
- Biederer T, Sara Y, Mozhayeva M, Atasoy D, Liu X, Kavalali ET, Südhof TC. SynCAM, a synaptic adhesion molecule that drives synapse assembly. Science. 2002;297:1525–1531. PubMed. - PubMed
- Chen L, Chetkovich DM, Petralia RS, Sweeney NT, Kawasaki Y, Wenthold RJ, Bredt DS, Nicoll RA. Stargazin regulates synaptic targeting of AMPA receptors by two distinct mechanisms. Nature. 2000;408:936–943. PubMed. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 NS067216/NS/NINDS NIH HHS/United States
- R21 NS052772/NS/NINDS NIH HHS/United States
- R21 NS052772-02/NS/NINDS NIH HHS/United States
- NS052772/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases