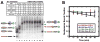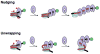Nucleosome remodeling by hMSH2-hMSH6 - PubMed (original) (raw)
Nucleosome remodeling by hMSH2-hMSH6
Sarah Javaid et al. Mol Cell. 2009.
Abstract
DNA nucleotide mismatches and lesions arise on chromosomes that are a complex assortment of protein and DNA (chromatin). The fundamental unit of chromatin is a nucleosome that contains approximately 146 bp DNA wrapped around an H2A, H2B, H3, and H4 histone octamer. We demonstrate that the mismatch recognition heterodimer hMSH2-hMSH6 disassembles a nucleosome. Disassembly requires a mismatch that provokes the formation of hMSH2-hMSH6 hydrolysis-independent sliding clamps, which translocate along the DNA to the nucleosome. The rate of disassembly is enhanced by actual or mimicked acetylation of histone H3 within the nucleosome entry-exit and dyad axis that occurs during replication and repair in vivo and reduces DNA-octamer affinity in vitro. Our results support a passive mechanism for chromatin remodeling whereby hMSH2-hMSH6 sliding clamps trap localized fluctuations in nucleosome positioning and/or wrapping that ultimately leads to disassembly, and highlight unanticipated strengths of the Molecular Switch Model for mismatch repair (MMR).
2009 Elsevier Inc.
Figures
Figure 1. Binding of hMSH2-hMSH6 to Nucleosome-DNA
(A) The Nucleosome-DNA substrate contains 17 bp 3′ of the 147 bp 5S rDNA nucleosome localization sequence (red) followed by a 28 bp linker, 24 bp lac O sequence (yellow), and 47 bp containing a mismatch site 20 bp from the 3′-end that contains a terminal biotin (light blue). (B) Representative gel showing specific binding of hMSH2-hMSH6 to the G/T mismatch nucleosome-DNA substrate containing an unmodified nucleosome. Boxes above indicate added reaction components (+), the concentration of hMSH2-hMSH6 (nM), and the inclusion of nucleosome-DNA (N). A schematic of DNA species with arrows or brackets indicating gel mobility position is shown on the left. The DNA substrate is colored as shown in (A) with a nucleosome (blue oval), hMSH2-hMSH6 (purple clamp); and streptavidin (green circle). (C) Quantitative analysis of hMSH2-hMSH6 binding to free-DNA containing a G/T mismatch (G/T) or G/C duplex (G/C) without or with biotin-streptavidin (-b*) on the 3′-end; and nucleosome-DNA with an unmodified (UN) or H3(K115Ac, K122Ac) modified (2Ac) nucleosome without or with (-b*). Standard deviations were determined from at least three independent experiments and error bars shown (some within the symbol).
Figure 2. Nucleosome Disassembly by hMSH2-hMSH6
Representative gels showing the nucleosome disassembly reaction catalyzed by hMSH2-hMSH6 with (A) G/T mismatch nucleosome-DNA containing an unmodified nucleosome, (B) G/T mismatch nucleosome-DNA containing an H3(K56Q) acetylation mimic nucleosome, and (C) G/T mismatch nucleosome-DNA containing an H3(K115Ac, K122Ac) modified nucleosome. Black bars indicate image splicing from a single gel where spliced out lanes were redundant with Fig. 3B. Boxes above indicate added reaction components (+) and the inclusion of free-DNA (F) or nucleosome-DNA (N). A schematic of DNA species with arrows or brackets indicating gel mobility position is shown on the left and right of the gel panels. The DNA substrate is colored as shown in Fig. 1A with a nucleosome (blue oval), hMSH2-hMSH6 (purple clamp); and streptavidin (green circle). Asterisks indicate the mobility of nucleosome-DNA substrate with bound hMSH2-hMSH6 and without a biotin-streptavidin bound 3′-tail. Red arrow indicates the gel mobility of the nucleosome disassembly product. Asterisk (*) indicates the position of the nucleosome substrate bound by hMSH2-hMSH6; multiple bands are consistent with multiple nucleosome positions surrounding the 5S rDNA localization site (see Suppl. Fig. 1). (D) Quantitative analysis of the nucleosome disassembly reactions. Data analysis includes representative gels shown in panel A and B as well as Supplemental Fig. 2. Each data set was fit to a single exponential decay to calculate τ and t1/2. Key: Unmodified nucleosome substrate containing duplex DNA (G/C) and biotin-streptavidin blocked (b*) 3′-tail (UN Nuc-G/C-b*); Unmodified nucleosome substrate containing a G/T mismatch and biotin-streptavidin blocked 3′-tail (UN Nuc-G/T-b*); H3(K56Q) acetylation mimic nucleosome substrate containing a G/T mismatch and biotin-streptavidin blocked 3′-tail (K56Q Nuc-G/T-b*); H3(K115Ac, K122Ac) nucleosome substrate containing duplex DNA (G/C) and biotin-streptavidin blocked 3′-tail (2Ac Nuc-G/C-b*); H3(K115Ac, K122Ac) nucleosome substrate containing a G/T mismatch and biotin-streptavidin blocked 3′-tail (2Ac Nuc-G/T-b*). Standard deviations were determined from at least three independent experiments and error bars shown (some within the symbol).
Figure 3. Analysis of the ATP Requirement for hMSH2-hMSH6 Nucleosome Disassembly
Boxes above indicate added reaction components (+), the inclusion of free-DNA (F) or Nucleosome-DNA (N), and the time of incubation (min). A schematic of DNA species with arrows or brackets indicating gel mobility position is shown on the left and right of the gel panels. The DNA substrate is colored as shown in Fig. 1A with a nucleosome (blue oval), hMSH2-hMSH6 (purple clamp); and streptavidin (green circle). Asterisks indicate the mobility of nucleosome-DNA substrate with bound hMSH2-hMSH6 and without a biotin-streptavidin bound 3′-tail. Red arrow indicates the gel mobility of the nucleosome disassembly product. (A) Nucleosome disassembly by hMSH2(K675A)-hMSH5(K1140A). Black bar indicates image splicing from a single gel where spliced lanes contained redundant controls shown in Fig. 2A, 2B, and 3B (lanes 6 and 7). (B) Nucleosome disassembly by hMSH2-hMSH6 in the presence of ATPγS. (C) and (D) Quantitative analysis of (A) plus Suppl. Fig. 4 A–C and (B) plus Suppl. Fig. 4 D–F, respectively. Each data set was fit to a single exponential decay to calculate and t1/2. See Fig. 2 for Key. Standard deviations were determined from at least three independent experiments and error bars shown (some within the symbol). (E) hMSH2-hMSH6 steady-state ATPase activity. hMSH2-hMSH6 ATPase activity was determined in the absence of DNA (no DNA), with free-DNA containing a G/T mismatch (G/T) or G/C duplex (G/C) with one (-b*) or two (*b-X-b*) biotin-streptavidin blocked ends, or with nucleosome-DNA containing an unmodified (UN-Nuc) or H3(K115Ac. K122Ac) modified (2Ac-Nuc) nucleosome and a G/T mismatch (G/T) or G/C duplex (G/C) without or with (-b*) a biotin-streptavidin blocked 3′-end. Standard deviations were determined from at least three independent experiments and error bars shown. A diagram of two ATPase cycles is shown on the right. Cycle A illustrates an ATPase cycle for free-DNA containing a single biotin-streptavidin blocked 3′-end (Gradia et al., 1999). Cycle B illustrates a hypothetical requirement for disassembly of a nucleosome from nucleosome-DNA containing a biotin-streptavidin blocked 3′-end to complete an ATPase cycle consistent with the data. The dashed blue arrow shows that the two cycles are connected by the product of nucleosome disassembly, which is identical to free-DNA containing a single biotin-streptavidin blocked 3′-end that initiates cycle A.
Figure 4. The Effect of Intervening Lac I on hMSH2-hMSH6 Nucleosome Disassembly
(A) Lac I blocks hMSH2-hMSH6 nucleosome disassembly. Black bar indicates image splicing from a single gel where spliced lanes contained redundant controls shown in Fig. 2A, 2B, and 3B (lanes 6 and 7). Boxes above indicate added reaction components (+), the inclusion of free-DNA (F) or Nucleosome-DNA (N), and the time of incubation (min). A schematic of DNA species with arrows or brackets indicating gel mobility position is shown on the left and right of the gel panels. The DNA substrate is colored as shown in Fig. 1A with a nucleosome (blue oval), hMSH2-hMSH6 (purple clamp); streptavidin (green circle); and Lac I (orange). Asterisks indicate the mobility of nucleosome-DNA substrate with bound hMSH2-hMSH6 and without a biotin-streptavidin bound 3′-tail. Red arrow indicates the gel mobility of the nucleosome disassembly product. Green arrows are a redundant control with Fig. 2B and indicate gel mobility of the nucleosome-DNA and the disassembly product following 60 min incubation without Lac I. (B) Quantitative analysis of (A) plus Suppl. Fig. 5 A–C. Each data set was fit to a single exponential decay to calculate τ and t1/2. See Fig. 2 for Key. Standard deviations were determined from at least three independent experiments and error bars shown (some within the symbol).
Figure 5. Two passive models for chromatin remodeling by hMSH2-hMSH6
Both models use the translocation of hMSH2-hMSH6 hydrolysis-independent sliding clamps to trap thermal fluctuations in the nucleosome structure. See text.
Similar articles
- Regulation of the nucleosome unwrapping rate controls DNA accessibility.
North JA, Shimko JC, Javaid S, Mooney AM, Shoffner MA, Rose SD, Bundschuh R, Fishel R, Ottesen JJ, Poirier MG. North JA, et al. Nucleic Acids Res. 2012 Nov 1;40(20):10215-27. doi: 10.1093/nar/gks747. Epub 2012 Sep 10. Nucleic Acids Res. 2012. PMID: 22965129 Free PMC article. - Phosphorylation of histone H3(T118) alters nucleosome dynamics and remodeling.
North JA, Javaid S, Ferdinand MB, Chatterjee N, Picking JW, Shoffner M, Nakkula RJ, Bartholomew B, Ottesen JJ, Fishel R, Poirier MG. North JA, et al. Nucleic Acids Res. 2011 Aug;39(15):6465-74. doi: 10.1093/nar/gkr304. Epub 2011 May 16. Nucleic Acids Res. 2011. PMID: 21576235 Free PMC article. - A quantitative model of nucleosome dynamics.
Forties RA, North JA, Javaid S, Tabbaa OP, Fishel R, Poirier MG, Bundschuh R. Forties RA, et al. Nucleic Acids Res. 2011 Oct;39(19):8306-13. doi: 10.1093/nar/gkr422. Epub 2011 Jul 14. Nucleic Acids Res. 2011. PMID: 21764779 Free PMC article. - All roads lead to chromatin: multiple pathways for histone deposition.
Li Q, Burgess R, Zhang Z. Li Q, et al. Biochim Biophys Acta. 2013 Mar-Apr;1819(3-4):238-46. Biochim Biophys Acta. 2013. PMID: 24459726 Review. - Mechanisms of ATP-dependent nucleosome sliding.
Bowman GD. Bowman GD. Curr Opin Struct Biol. 2010 Feb;20(1):73-81. doi: 10.1016/j.sbi.2009.12.002. Epub 2010 Jan 8. Curr Opin Struct Biol. 2010. PMID: 20060707 Free PMC article. Review.
Cited by
- Nucleosome DNA unwrapping does not affect prototype foamy virus integration efficiency or site selection.
Mackler RM, Jones ND, Gardner AM, Lopez MA Jr, Howard CJ, Fishel R, Yoder KE. Mackler RM, et al. PLoS One. 2019 Mar 13;14(3):e0212764. doi: 10.1371/journal.pone.0212764. eCollection 2019. PLoS One. 2019. PMID: 30865665 Free PMC article. - Preparation of fully synthetic histone H3 reveals that acetyl-lysine 56 facilitates protein binding within nucleosomes.
Shimko JC, North JA, Bruns AN, Poirier MG, Ottesen JJ. Shimko JC, et al. J Mol Biol. 2011 Apr 29;408(2):187-204. doi: 10.1016/j.jmb.2011.01.003. Epub 2011 Feb 15. J Mol Biol. 2011. PMID: 21310161 Free PMC article. - Regulation of the nucleosome unwrapping rate controls DNA accessibility.
North JA, Shimko JC, Javaid S, Mooney AM, Shoffner MA, Rose SD, Bundschuh R, Fishel R, Ottesen JJ, Poirier MG. North JA, et al. Nucleic Acids Res. 2012 Nov 1;40(20):10215-27. doi: 10.1093/nar/gks747. Epub 2012 Sep 10. Nucleic Acids Res. 2012. PMID: 22965129 Free PMC article. - Overview for the histone codes for DNA repair.
Williamson EA, Wray JW, Bansal P, Hromas R. Williamson EA, et al. Prog Mol Biol Transl Sci. 2012;110:207-27. doi: 10.1016/B978-0-12-387665-2.00008-0. Prog Mol Biol Transl Sci. 2012. PMID: 22749147 Free PMC article. Review. - Assessment of anti-recombination and double-strand break-induced gene conversion in human cells by a chromosomal reporter.
Xu K, Wu X, Tompkins JD, Her C. Xu K, et al. J Biol Chem. 2012 Aug 24;287(35):29543-53. doi: 10.1074/jbc.M112.352302. Epub 2012 Jul 7. J Biol Chem. 2012. PMID: 22773873 Free PMC article.
References
- Acharya S, Foster PL, Brooks P, Fishel R. The coordinated functions of the E. coli MutS and MutL proteins in mismatch repair. Molecular Cell. 2003;12:233–246. - PubMed
- Ataian Y, Krebs JE. Five repair pathways in one context: chromatin modification during DNA repair. Biochem Cell Biol. 2006;84:490–504. - PubMed
- Boland CR, Fishel R. Lynch syndrome: form, function, proteins, and basketball. Gastroenterology. 2005;129:751–755. - PubMed
- Drummond JT, Li GM, Longley MJ, Modrich P. Isolation of an hMSH2-p160 heterodimer that restores DNA mismatch repair to tumor cells [see comments] Science. 1995;268:1909–1912. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM080176/GM/NIGMS NIH HHS/United States
- R01 GM083055/GM/NIGMS NIH HHS/United States
- R01 GM062556/GM/NIGMS NIH HHS/United States
- CA067007/CA/NCI NIH HHS/United States
- R01 CA067007/CA/NCI NIH HHS/United States
- GM062556/GM/NIGMS NIH HHS/United States
- R01 CA067007-16/CA/NCI NIH HHS/United States
- R01 GM080176-17S1/GM/NIGMS NIH HHS/United States
- GM083055/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources