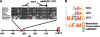Upregulation of c-MYC in cis through a large chromatin loop linked to a cancer risk-associated single-nucleotide polymorphism in colorectal cancer cells - PubMed (original) (raw)
Upregulation of c-MYC in cis through a large chromatin loop linked to a cancer risk-associated single-nucleotide polymorphism in colorectal cancer cells
Jason B Wright et al. Mol Cell Biol. 2010 Mar.
Abstract
Genome-wide association studies have mapped many single-nucleotide polymorphisms (SNPs) that are linked to cancer risk, but the mechanism by which most SNPs promote cancer remains undefined. The rs6983267 SNP at 8q24 has been associated with many cancers, yet the SNP falls 335 kb from the nearest gene, c-MYC. We show that the beta-catenin-TCF4 transcription factor complex binds preferentially to the cancer risk-associated rs6983267(G) allele in colon cancer cells. We also show that the rs6983267 SNP has enhancer-related histone marks and can form a 335-kb chromatin loop to interact with the c-MYC promoter. Finally, we show that the SNP has no effect on the efficiency of chromatin looping to the c-MYC promoter but that the cancer risk-associated SNP enhances the expression of the linked c-MYC allele. Thus, cancer risk is a direct consequence of elevated c-MYC expression from increased distal enhancer activity and not from reorganization/creation of the large chromatin loop. The findings of these studies support a mechanism for intergenic SNPs that can promote cancer through the regulation of distal genes by utilizing preexisting large chromatin loops.
Figures
FIG. 1.
The rs6983267 SNP is highly conserved in mammalian evolution and maps to a consensus TCF/LEF1 binding site. (A) Comparative genomics analysis of the rs6983267 SNP regions in the indicated mammals, including marsupials and monotremes. The ancestral element has a G, whereas the allelic frequency in humans is 50% T. Consensus inverted TCF binding sites are indicated by brackets. rs6983267 falls ∼335 kb in the 5′ direction from c-MYC in all mammals. (B) Alignment of the three top-scoring consensus TCF/LEF1 motifs (dTCF, Drosophila T-cell factor) is shown above the mammalian conserved sequence surrounding the rs6983267 SNP. Both the ancestral G variant, which is strongly associated with several cancers, and the non-cancer-associated T variant of rs6983267 are depicted (the variations are indicated by the rectangular black box).
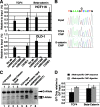
FIG. 2.
Enhanced binding of TCF4 and beta-catenin to the cancer-associated rs6983267 SNP in vivo. (A) ChIP analysis of TCF4 and beta-catenin binding in CRC cell lines HCT116 and DLD-1. Samples from independent, duplicate immunoprecipitations were assayed for the presence of three different cancer-associated SNPs: rs6983267 (associated with colon cancer and others), rs1447295 (associated with prostate cancer), and rs6942880 (associated with bladder cancer). A TCF4 binding site near the CCND1 gene was used as a positive control. ChIP allowed quantitation of TCF4 and beta-catenin binding to rs6983267 and other SNPs. The ChIP signal (expressed as a percentage relative to the input level) is shown for three different SNPs, as well as for the control TCF4 site in the CCND1 gene. No ab, no antibody. (B) The rs6983267 SNP region from DLD-1 cells was PCR amplified from either input DNA or chromatin immunoprecipitates obtained using anti-TCF or anti-beta-catenin antibodies. The PCR products were subjected to direct DNA sequencing using a primer from one end. The input DNA is heterozygous for rs6983267, whereas the ChIP DNA samples obtained with both TCF4 and beta-catenin antibodies show enrichment with the cancer-associated rs6983267(G) allele. (C) Restriction enzyme analysis of allele distribution in DLD-1 input DNA compared to that in ChIP DNA by using anti-TCF4 and anti-beta-catenin. Tsp45I cleaves the rs6983267(T) allele but not the rs6983267(G) allele. rs6983267(T)-only and rs6983267(G)-only amplification products were used to verify specificity and complete enzyme digestion (lanes 7 and 8). Independent duplicate ChIP samples were amplified for each antibody (lanes 1 to 4), and equivalent amounts of DNA were digested and loaded into the lanes. Equivalent samples of input DNA for the individual immunoprecipitations were also digested and loaded (lanes 5 and 6). (D) Quantitation of rs6983267 allelic distributions in input and ChIP samples by either direct sequencing or Tsp45I digestion. For each antibody, the peak height of the G/T allele ratio for the input DNA was set to 1 and used to normalize the G/T allele ratio for the ChIP DNA. All DNA samples were amplified in the same number of cycles and to the minimum extent needed for direct sequencing or digestion.
FIG. 3.
Allele-specific enhancer histone modifications at the cancer-associated rs6983267 SNP. (A) Quantitation of three different enhancer-associated histone modifications (H3K4me2, H3-Ac, and H4-Ac) at rs6983267, rs1447295, and rs6942880, as well as the region surrounding the TCF4 binding site in CCND1, by ChIP. Data are presented as percentages of the input level. (B) Allele specificity of histone modifications as determined by direct sequencing of the PCR products.
FIG. 4.
A 335-kb chromatin loop links the cancer-associated rs6983267 SNP to the c-MYC promoter. (A to D) 3C analyses of the c-MYC promoters in DLD-1 CRC cells, HCT116 CRC cells, MCF7 breast cancer cells, and K562 myeloid leukemia cells. Relative frequencies of interaction between primers derived from the 5′ flanking domain and the c-MYC promoter are shown. The data in panels A, B, and C are derived from three separate 3C analyses for each primer combination, and those in panel D are from one experiment. PCR products were normalized relative to positive-control templates. An interaction of the c-MYC promoter with rs6983267 but not with two other cancer-associated SNPs or other primers in the region was observed. (E) 3C analysis of the rs6983267 enhancer in DLD-1 cells. A primer from rs6983267 was combined with primers from POU5FP1, rs1447295, c-MYC, or PVT1. Relative interaction frequencies were calculated as described above.
FIG. 5.
Chromatin looping is not influenced by the rs6983267 SNP. The allele specificity of intrachromosomal looping was evaluated by sequencing of the 3C PCR product from an rs6983267/c-MYC primer pair. The two rs6983267 alleles are detected equally in both DLD-1 colon cancer cells and MCF7 breast cancer cells.
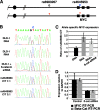
FIG. 6.
The cancer risk-associated rs6983267(G) allele is linked to enhanced c-MYC expression. (A) Diagram of the linked polymorphisms at c-MYC SNPs rs6983267 and rs4645953 in DLD-1 cells as determined by 3C analysis. (B) Allele-specific c-MYC expression as evaluated by PCR analysis of the c-MYC rs4645953 SNP in DLD-1 cells. Genomic DNA exhibits heterozygosity at the rs4645953 SNP (DLD-1 DNA profile). RT-PCR detects twofold-enhanced expression of the c-MYC rs4645953(C) allele (DLD-1 RNA profile). RT-PCR detects the same twofold-enhanced expression of the c-MYC rs4645953(C) allele after knockdown of beta-catenin expression with siRNA (DLD-1 + β-catenin siRNA profile). Sequencing profiles accurately detect allele-specific expression because transient transfection with cloned full-length c-MYC genes carrying the different rs4645953 alleles (C and T alleles) gives a 1:1 ratio of expression levels when the c-MYC genes are mixed in a 1:1 ratio (rs4645953 C/T 1:1 profile). Mixing the c-MYC rs4645953 C and T alleles in a 2:1 ratio gives a corresponding 2:1 ratio of expression levels as determined by RT-PCR (rs4645953 C/T 2:1 profile). (C) RT-PCR quantitation of c-MYC allele-specific expression in DLD-1 cells before and after knockdown of beta-catenin (β-cat) expression by using siRNA. (D) Knockdown of beta-catenin expression gives a corresponding reduction in c-MYC expression. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Similar articles
- A dynamic exchange of TCF3 and TCF4 transcription factors controls MYC expression in colorectal cancer cells.
Shah M, Rennoll SA, Raup-Konsavage WM, Yochum GS. Shah M, et al. Cell Cycle. 2015;14(3):323-32. doi: 10.4161/15384101.2014.980643. Cell Cycle. 2015. PMID: 25659031 Free PMC article. - The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling.
Tuupanen S, Turunen M, Lehtonen R, Hallikas O, Vanharanta S, Kivioja T, Björklund M, Wei G, Yan J, Niittymäki I, Mecklin JP, Järvinen H, Ristimäki A, Di-Bernardo M, East P, Carvajal-Carmona L, Houlston RS, Tomlinson I, Palin K, Ukkonen E, Karhu A, Taipale J, Aaltonen LA. Tuupanen S, et al. Nat Genet. 2009 Aug;41(8):885-90. doi: 10.1038/ng.406. Epub 2009 Jun 28. Nat Genet. 2009. PMID: 19561604 - VDR/RXR and TCF4/β-catenin cistromes in colonic cells of colorectal tumor origin: impact on c-FOS and c-MYC gene expression.
Meyer MB, Goetsch PD, Pike JW. Meyer MB, et al. Mol Endocrinol. 2012 Jan;26(1):37-51. doi: 10.1210/me.2011-1109. Epub 2011 Nov 22. Mol Endocrinol. 2012. PMID: 22108803 Free PMC article. - MYC association with cancer risk and a new model of MYC-mediated repression.
Cole MD. Cole MD. Cold Spring Harb Perspect Med. 2014 Jul 1;4(7):a014316. doi: 10.1101/cshperspect.a014316. Cold Spring Harb Perspect Med. 2014. PMID: 24985129 Free PMC article. Review. - The MYC Enhancer-ome: Long-Range Transcriptional Regulation of MYC in Cancer.
Lancho O, Herranz D. Lancho O, et al. Trends Cancer. 2018 Dec;4(12):810-822. doi: 10.1016/j.trecan.2018.10.003. Epub 2018 Nov 2. Trends Cancer. 2018. PMID: 30470303 Free PMC article. Review.
Cited by
- The many faces and functions of β-catenin.
Valenta T, Hausmann G, Basler K. Valenta T, et al. EMBO J. 2012 Jun 13;31(12):2714-36. doi: 10.1038/emboj.2012.150. Epub 2012 May 22. EMBO J. 2012. PMID: 22617422 Free PMC article. Review. - Melatonin and cancer suppression: insights into its effects on DNA methylation.
Davoodvandi A, Nikfar B, Reiter RJ, Asemi Z. Davoodvandi A, et al. Cell Mol Biol Lett. 2022 Sep 5;27(1):73. doi: 10.1186/s11658-022-00375-z. Cell Mol Biol Lett. 2022. PMID: 36064311 Free PMC article. - Cohesin is required for activation of MYC by estradiol.
McEwan MV, Eccles MR, Horsfield JA. McEwan MV, et al. PLoS One. 2012;7(11):e49160. doi: 10.1371/journal.pone.0049160. Epub 2012 Nov 8. PLoS One. 2012. PMID: 23145106 Free PMC article. - Inauhzin(c) inactivates c-Myc independently of p53.
Jung JH, Liao JM, Zhang Q, Zeng S, Nguyen D, Hao Q, Zhou X, Cao B, Kim SH, Lu H. Jung JH, et al. Cancer Biol Ther. 2015;16(3):412-9. doi: 10.1080/15384047.2014.1002698. Cancer Biol Ther. 2015. PMID: 25692307 Free PMC article. - Genetic heterogeneity in colorectal cancer associations between African and European americans.
Kupfer SS, Anderson JR, Hooker S, Skol A, Kittles RA, Keku TO, Sandler RS, Ellis NA. Kupfer SS, et al. Gastroenterology. 2010 Nov;139(5):1677-85, 1685.e1-8. doi: 10.1053/j.gastro.2010.07.038. Epub 2010 Jul 24. Gastroenterology. 2010. PMID: 20659471 Free PMC article.
References
- Amundadottir, L. T., P. Sulem, J. Gudmundsson, A. Helgason, A. Baker, B. A. Agnarsson, A. Sigurdsson, K. R. Benediktsdottir, J. B. Cazier, J. Sainz, M. Jakobsdottir, J. Kostic, D. N. Magnusdottir, S. Ghosh, K. Agnarsson, B. Birgisdottir, L. Le Roux, A. Olafsdottir, T. Blondal, M. Andresdottir, O. S. Gretarsdottir, J. T. Bergthorsson, D. Gudbjartsson, A. Gylfason, G. Thorleifsson, A. Manolescu, K. Kristjansson, G. Geirsson, H. Isaksson, J. Douglas, J. E. Johansson, K. Balter, F. Wiklund, J. E. Montie, X. Yu, B. K. Suarez, C. Ober, K. A. Cooney, H. Gronberg, W. J. Catalona, G. V. Einarsson, R. B. Barkardottir, J. R. Gulcher, A. Kong, U. Thorsteinsdottir, and K. Stefansson. 2006. A common variant associated with prostate cancer in European and African populations. Nat. Genet. 38:652-658. - PubMed
- Bailey, T. L., and C. Elkan. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2:28-36. - PubMed
- Cesarman, E., R. Dalla-Favera, D. Bentley, and M. Groudine. 1987. Mutations in the first exon are associated with altered transcription of c-myc in Burkitt lymphoma. Science 238:1272-1275. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical