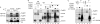The DNA unwinding element binding protein DUE-B interacts with Cdc45 in preinitiation complex formation - PubMed (original) (raw)
The DNA unwinding element binding protein DUE-B interacts with Cdc45 in preinitiation complex formation
A Chowdhury et al. Mol Cell Biol. 2010 Mar.
Abstract
Template unwinding during DNA replication initiation requires the loading of the MCM helicase activator Cdc45 at replication origins. We show that Cdc45 interacts with the DNA unwinding element (DUE) binding protein DUE-B and that these proteins localize to the DUEs of active replication origins. DUE-B and Cdc45 are not bound at the inactive c-myc replicator in the absence of a functional DUE or at the recently identified ataxin 10 (ATX10) origin, which is silent before disease-related (ATTCT)(n) repeat length expansion of its DUE sequence, despite the presence of the origin recognition complex (ORC) and MCM proteins at these origins. Addition of a heterologous DUE to the ectopic c-myc origin, or expansion of the ATX10 DUE, leads to origin activation, DUE-B binding, and Cdc45 binding. DUE-B, Cdc45, and topoisomerase IIbeta binding protein 1 (TopBP1) form complexes in cell extracts and when expressed from baculovirus vectors. During replication in Xenopus egg extracts, DUE-B and Cdc45 bind to chromatin with similar kinetics, and DUE-B immunodepletion blocks replication and the loading of Cdc45 and a fraction of TopBP1. The coordinated binding of DUE-B and Cdc45 to origins and the physical interactions of DUE-B, Cdc45, and TopBP1 suggest that complexes of these proteins are necessary for replication initiation.
Figures
FIG. 1.
Protein binding to replication origins. (A) DUE-B (i) and Cdc45 (ii) binding to the c-myc and lamin B2 origins (ori) in HeLa cells and VM (SCA10 patient) cells. Maps show the positions of sequence-tagged sites (STS) used for quantitative ChIP within the c-myc and lamin B2 origins. Diamond shapes, DUEs predicted by the SIDD (stress-induced DNA duplex destabilization) algorithm (3, 7). Restriction sites flanking the c-myc replicator core: H, HindIII; X, XhoI. (B) Origin activity (nascent strand abundance) (i) and Orc2 (ii), Mcm4 (iii), DUE-B (iv), Cdc45 (v), and RPA (vi) binding at the ATX10/SCA10 locus (map at bottom) in non-SCA10 lymphoblastoid cells (left, 428-12) and SCA10 patient cells (right, VM). Diamond shape, position of expanded (ATTCT)n repeats in SCA10 patient cells. (C) Origin activity (i) and Orc2, Mcm3, DUE-B, and Cdc45 binding (ii) at the ectopic c-myc replicator (map at bottom, STS pUV) in FLP-RMCE cell lines containing the wild-type (WT; 406.myc) c-myc replicator, the c-myc replicator minus the DUE (ΔDUE), or the c-myc replicator in which the DUE has been replaced by the SCA10 DUE containing (ATTCT)8, (ATTCT)13, (ATTCT)27, or (ATTCT)48 repeats. The data shown in panels Bi and Ci are reproduced from reference (copyright, American Society for Microbiology; reproduced with permission).
FIG. 2.
Replication protein abundance and chromatin binding. (A, left) Whole-cell extract (WCE) levels of Orc2, DUE-B, and β-actin; (right) chromatin-bound Orc3, Mcm7, and DUE-B in HCT116 cells or HCT116 Orc2Δ/− cells. Ku80 was used as a loading control (20, 38). (B) DUE-B, β-actin, and FHIT protein levels were assayed in permeabilized nuclei (lanes 1 to 4) and WCE (lanes 5 to 8) after serum (0.5%) deprivation of cells for 0, 1, and 3 days (lanes 1 to 3 and 5 to 7) and after 3 days of serum deprivation, followed by 3 days of serum (10%) restoration (lanes 4 and 8). (C) The levels of the indicated proteins were compared in WCE from control (scrambled) siRNA- or DUE-B siRNA-treated HeLa cells. (D and E) Chromatin binding of the indicated proteins in control siRNA- or DUE-B siRNA-treated HeLa cells.
FIG. 3.
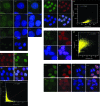
Indirect immunofluorescence localization of DUE-B. (A) HeLa cells stained with anti-DUE-B antibody (A1) and DAPI (A2) (merged image [A3]). (B) HeLa A1 cells expressing epitope (His6)-tagged DUE-B stained with anti-His6 antibody (B1) and DAPI (B2) (merged image [B3]). (C) Staining done as in panel B, with 0.2% Triton X-100 preextraction. (D) Negative-control HeLa cells stained with anti-His6 antibody (D1) and DAPI (D2). (E) HeLa A1 cells pulse-labeled with BrUdR and stained with anti-His6 antibody (E1), anti-BrUdR antibody (E2), and DAPI (E3) (merged image [E4]). (E5) Plot showing degree of colocalization of DUE-B (λ = 617 nm) and BrUdR (λ = 528 nm) signals (Pearson correlation coefficient, ρ = 0.4084). (F) HeLa A1 cells stained with anti-His6 antibody (F1), anti-SAP145 antibody (F2), and DAPI (F3) (merged image [F4]). (F5) Plot showing degree of colocalization of DUE-B (λ = 528 nm) and SAP145 (λ = 617 nm) signals (Pearson correlation coefficient, ρ = 0.7680). (G) HeLa A1 cells stained with anti-His6 antibody (G1), anti-SM antibody (G2), and DAPI (G3) (merged image [G4]). (G5) Plot showing degree of colocalization of DUE-B (λ = 528 nm) and Sm (λ = 617 nm) signals (Pearson correlation coefficient, ρ = 0.6492). (H) HeLa A1 cells stained with anti-His6 antibody (H1, H5), anti-SAP145 antibody (H2, H6), and DAPI (H3, H7) (merged images [H4, H8]). Arrowheads indicate cells in mitosis.
FIG. 4.
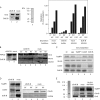
Protein binding to chromatin-Xenopus cell-free replication system. (A) Time course of binding of the indicated proteins to sperm chromatin. *, cross-reactive band. (B) Time course of protein binding to bead-bound plasmid DNA. (C) Geminin inhibits Mcm7, DUE-B, and Cdc45 binding to sperm chromatin. Lane 1, egg extract (xtr); lanes 2 to 5, control (no geminin); lane 6, control (sperm chromatin omitted); lane 7, egg extract (geminin added); lanes 8 to 11, geminin treated. (D) Inhibition of DUE-B and Cdc45 binding to sperm chromatin (chr) by roscovitine. Lanes 1 and 2, untreated; lanes 4 and 5, roscovitine treated. Lanes 1 and 5, protein bound to sperm chromatin (45′ time point); lanes 2 and 4, egg extract. Lane 3, protein markers. (E) Inhibition of DUE-B and Cdc45 binding to sperm chromatin by p27Kip. Lanes 1 to 3, untreated; lanes 4 to 6, p27Kip treated.
FIG. 5.
Interaction of DUE-B, Cdc45, and TopBP1. (A) Immunoprecipitation (IP) of HeLa cell extracts using normal rabbit serum (NRS, lane 1) or antibody against DUE-B (lane 2), Cdc45 (lane 3), or TopBP1 (lane 4). Proteins indicated to the right of the figure were visualized by Western blotting (WB) with the cognate antibodies. (B) Western blot of lysates (input) of Sf9 cells infected pairwise with baculoviruses expressing DUE-B, TopBP1, and Cdc45 (lanes 1 to 3, unfilled ovals). Lane 4, IP with preimmune IgG; lanes 5 to 10, IP with antibody (filled ovals) against one of each pair of baculovirus-expressed proteins. (C) Western blot of immunoprecipitates of lysates (input) of Sf9 cells infected pairwise with baculoviruses expressing ΔCT DUE-B, TopBP1, and Cdc45 (lanes 1 and 4, unfilled ovals). Lanes 2, 3, 5, and 6, IP with antibody (filled ovals) against one of each pair of baculovirus-expressed proteins.
FIG. 6.
Alignment of yeast Sld3 and DUE-B. (A to C) The carboxy-terminal regions of Sld3 from S. cerevisiae Sld3 (ScSld3), S. pombe Sld3 (SpSld3), and human DUE-B were aligned as indicated, using DNAMAN. Amino acid similarity (47) and identity are indicated by light and dark outlines, respectively. (D) CK2 consensus target sites (numbered) in DUE-B.
FIG. 7.
Phosphorylation of DUE-B. (A) Recombinant DUE-B was purified by Ni-NTA chromatography from HeLa A1 cells and visualized by antiphosphoserine antibody before (−) or after (+) treatment with λ phosphatase (PP'ase). (B) Epitope-tagged (His6) full-length DUE-B or the ΔCT-DUE-B fragment was expressed in baculovirus-infected Sf9 cells and purified. Proteins were untreated (lanes 1 and 3) or treated (lanes 2 and 4) with casein kinase 2 (CK2) and [γ-32P]ATP. Top, autoradiogram; bottom, anti-His6 Western blot. (C) Baculovirus-expressed DUE-B was phosphorylated by CK2 in the presence of [γ-32P]ATP and electrophoresed before or after treatment with formic acid, which cleaves preferentially between D156 and P157 (18). Top, autoradiogram; bottom, anti-DUE-B Western blot. NT, N-terminal fragment; CT, C-terminal fragment. (D) Egg extracts were either untreated (lanes 1, 4, and 7), added to baculovirus-expressed His6-tagged DUE-B that was untreated (lanes 2, 5, and 8), or phosphorylated by CK2 (lanes 3, 6, and 9). Ni-NTA beads were added to each extract and washed with buffer of the indicated NaCl concentration, and the proteins pulled down were visualized by Western blotting with anti-DUE-B, anti-Cdc45, or anti-TopBP1 antibodies.
FIG. 8.
DUE-B-dependent loading of Cdc45. (A) Xenopus egg extract was immunodepleted (left) with anti-DUE-B antibody (lane 1, ΔDUE-B) or preimmune serum (lane 2, mock), and buffer or recombinant His6-DUE-B purified from HeLa A1 cells (lane 3, Coomassie-stained gel) was added to the extracts for time course analysis of sperm chromatin replication (right). (B) Sperm chromatin was added to extract immunodepleted with anti-DUE-B antibody (lane 1) or preimmune serum (lane 2, mock) for analysis of the time course of Orc2 and Mcm3 binding (lanes 3 to 8). (C) Xenopus egg extract was undepleted (lane 1) or immunodepleted with preimmune serum (lane 2) or antiserum against TopBP1 (lane 3), Cdc45 (lane 4), or DUE-B (lane 5). (D) Extract was immunodepleted with anti-DUE-B antibody (lane 1) or preimmune serum (lane 2), and sperm chromatin was added for analysis of the time course of Cdc45 binding (lanes 3 to 6). The loading control is a nonspecific cross-reacting band. (E) Lane 1, His6-DUE-B purified from HeLa A1 cells for add back (silver-stained gel). Lanes 2 to 4, TopBP1, Cdc45, and Orc2 binding to sperm chromatin (45′ time point) in DUE-B-immunodepleted extract (lane 2), mock-depleted extract (lane 3), or DUE-B-immunodepleted extract containing His6-DUE-B purified from HeLa A1 cells (lane 4).
FIG. 9.
Model for pre-IC formation. In this model, interacting DUE-B, Cdc45, and TopBP1 associate with the pre-RC. Cdk2 phosphorylation triggers Cdc45 activation of the pre-IC. Upon origin unwinding, DUE-B is released, and RPA binds to the origin, followed by replisome assembly and DNA synthesis. See the text for further details.
Similar articles
- Interaction between DUE-B and Treslin is required to load Cdc45 on chromatin in human cells.
Poudel S, Yao J, Kemp MG, Leffak M. Poudel S, et al. J Biol Chem. 2018 Sep 14;293(37):14497-14506. doi: 10.1074/jbc.RA118.004519. Epub 2018 Jul 23. J Biol Chem. 2018. PMID: 30037903 Free PMC article. - MCM2-7 complexes bind chromatin in a distributed pattern surrounding the origin recognition complex in Xenopus egg extracts.
Edwards MC, Tutter AV, Cvetic C, Gilbert CH, Prokhorova TA, Walter JC. Edwards MC, et al. J Biol Chem. 2002 Sep 6;277(36):33049-57. doi: 10.1074/jbc.M204438200. Epub 2002 Jun 26. J Biol Chem. 2002. PMID: 12087101 - Protein phosphatase 2A and Cdc7 kinase regulate the DNA unwinding element-binding protein in replication initiation.
Gao Y, Yao J, Poudel S, Romer E, Abu-Niaaj L, Leffak M. Gao Y, et al. J Biol Chem. 2014 Dec 26;289(52):35987-6000. doi: 10.1074/jbc.M114.589119. Epub 2014 Sep 25. J Biol Chem. 2014. PMID: 25258324 Free PMC article. - Behavior of replication origins in Eukaryota - spatio-temporal dynamics of licensing and firing.
Musiałek MW, Rybaczek D. Musiałek MW, et al. Cell Cycle. 2015;14(14):2251-64. doi: 10.1080/15384101.2015.1056421. Epub 2015 Jun 1. Cell Cycle. 2015. PMID: 26030591 Free PMC article. Review. - Eukaryotic DNA replication: from pre-replication complex to initiation complex.
Takisawa H, Mimura S, Kubota Y. Takisawa H, et al. Curr Opin Cell Biol. 2000 Dec;12(6):690-6. doi: 10.1016/s0955-0674(00)00153-8. Curr Opin Cell Biol. 2000. PMID: 11063933 Review.
Cited by
- GEMC1 is a novel TopBP1-interacting protein involved in chromosomal DNA replication.
Piergiovanni G, Costanzo V. Piergiovanni G, et al. Cell Cycle. 2010 Sep 15;9(18):3662-6. doi: 10.4161/cc.9.18.13060. Epub 2010 Sep 16. Cell Cycle. 2010. PMID: 20855966 Free PMC article. - Protein phosphatases in chromatin structure and function.
Gil RS, Vagnarelli P. Gil RS, et al. Biochim Biophys Acta Mol Cell Res. 2019 Jan;1866(1):90-101. doi: 10.1016/j.bbamcr.2018.07.016. Epub 2018 Jul 20. Biochim Biophys Acta Mol Cell Res. 2019. PMID: 30036566 Free PMC article. Review. - Non-B DB: a database of predicted non-B DNA-forming motifs in mammalian genomes.
Cer RZ, Bruce KH, Mudunuri US, Yi M, Volfovsky N, Luke BT, Bacolla A, Collins JR, Stephens RM. Cer RZ, et al. Nucleic Acids Res. 2011 Jan;39(Database issue):D383-91. doi: 10.1093/nar/gkq1170. Epub 2010 Nov 21. Nucleic Acids Res. 2011. PMID: 21097885 Free PMC article. - Order from clutter: selective interactions at mammalian replication origins.
Aladjem MI, Redon CE. Aladjem MI, et al. Nat Rev Genet. 2017 Feb;18(2):101-116. doi: 10.1038/nrg.2016.141. Epub 2016 Nov 21. Nat Rev Genet. 2017. PMID: 27867195 Free PMC article. Review. - Regulating DNA replication in eukarya.
Siddiqui K, On KF, Diffley JF. Siddiqui K, et al. Cold Spring Harb Perspect Biol. 2013 Sep 1;5(9):a012930. doi: 10.1101/cshperspect.a012930. Cold Spring Harb Perspect Biol. 2013. PMID: 23838438 Free PMC article. Review.
References
- Aggarwal, B. D., and B. R. Calvi. 2004. Chromatin regulates origin activity in Drosophila follicle cells. Nature 430:372-376. - PubMed
- Aparicio, O. M., D. M. Weinstein, and S. P. Bell. 1997. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell 91:59-69. - PubMed
- Bell, S. P., and B. Stillman. 1992. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature 357:128-134. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM053819/GM/NIGMS NIH HHS/United States
- R15 GM084407/GM/NIGMS NIH HHS/United States
- GM82995/GM/NIGMS NIH HHS/United States
- R01 GM082995/GM/NIGMS NIH HHS/United States
- GM53819/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous