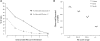Evolutionary constraint facilitates interpretation of genetic variation in resequenced human genomes - PubMed (original) (raw)
Evolutionary constraint facilitates interpretation of genetic variation in resequenced human genomes
David L Goode et al. Genome Res. 2010 Mar.
Abstract
Here, we demonstrate how comparative sequence analysis facilitates genome-wide base-pair-level interpretation of individual genetic variation and address two questions of importance for human personal genomics: first, whether an individual's functional variation comes mostly from noncoding or coding polymorphisms; and, second, whether population-specific or globally-present polymorphisms contribute more to functional variation in any given individual. Neither has been definitively answered by analyses of existing variation data because of a focus on coding polymorphisms, ascertainment biases in favor of common variation, and a lack of base-pair-level resolution for identifying functional variants. We resequenced 575 amplicons within 432 individuals at genomic sites enriched for evolutionary constraint and also analyzed variation within three published human genomes. We find that single-site measures of evolutionary constraint derived from mammalian multiple sequence alignments are strongly predictive of reductions in modern-day genetic diversity across a range of annotation categories and across the allele frequency spectrum from rare (<1%) to high frequency (>10% minor allele frequency). Furthermore, we show that putatively functional variation in an individual genome is dominated by polymorphisms that do not change protein sequence and that originate from our shared ancestral population and commonly segregate in human populations. These observations show that common, noncoding alleles contribute substantially to human phenotypes and that constraint-based analyses will be of value to identify phenotypically relevant variants in individual genomes.
Figures
Figure 1.
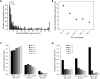
Derived allele frequency (DAF) compared with evolutionary constraint. (A) DAF spectrum of all SNVs. Each category is one percentile DAF. Note the much higher number of SNVs with 0%–1% and 1%–2% DAF compared with the rest of the data. (B) Mean DAF as a function of RS score. Sites with RS > 0 are binned by increasing level of constraint. (C) Proportion of sites of the indicated DAF within each of five RS bins. Bars of each RS bin add up to 1 but are organized by DAF to facilitate visual comparison between the RS bins. The greater the RS of a site, the rarer is its derived allele. (D) Proportion of sites within the indicated RS bins at sites of rare, intermediate to common, and very common DAF. The greater the DAF, the more SNVs avoid constrained sites.
Figure 2.
Variation and evolutionary constraint in three personal genomes. (A) Coverage of the human genome by mammalian alignment depth (solid line) and level of constraint (broken line). (B) Ratio of the number of SNVs observed in the three individual genomes at sites within the given RS score range to the number expected, given the distribution of RS scores across the human genome.
Figure 3.
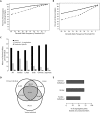
Properties of derived alleles at constrained sites. (A) Percentage of sites homozygous for derived alleles, where the derived alleles are of a frequency above the threshold indicated along the _x_-axis, per individual, at all sites (solid line) or at constrained sites only (broken lines). (B) Percentage of derived alleles of a frequency above the threshold indicated along the _x_-axis, per individual, at all sites (solid line) or at constrained sites only (broken lines). (C) Mean number of sites per individual that bear derived alleles found in all five populations (global SNVs), are found only in the indicated population (population specific), or are shared between two to four populations. (D) Number of segregating sites in each individual at highly constrained (RS > 2) sites that are shared between individuals or are private to each individual. (E) Percentage of segregating sites at constrained (RS > 2) sites in each individual that are shared with at least one of the other individuals.
Figure 4.
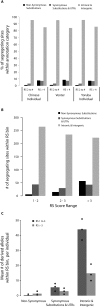
Relative abundance of coding and noncoding functional variation. SNVs were divided into three categories: those that cause nonsynonymous substitutions; those that cause synonymous substitutions or changes in the UTRs; and those that do not occur in exons (intronic and intergenic). (A) Contribution of SNVs in each category to total variation at constrained sites in each resequenced individual genome at sites with RS 2 to 4 and at sites with RS > 4. (B) Total number of segregating sites in our ENCODE resequencing sample that occur at constrained sites in our three annotation categories. Constrained sites are divided into bins of increasing constraint. (C) Mean number of segregating sites carried by the individuals in our ENCODE resequencing sample in all three categories, at moderately (RS 1 to 3) and highly (RS > 3) constrained positions. Diamonds correspond to the 10%, 50%, and 90% quantiles.
Similar articles
- Analysis of sequence conservation at nucleotide resolution.
Asthana S, Roytberg M, Stamatoyannopoulos J, Sunyaev S. Asthana S, et al. PLoS Comput Biol. 2007 Dec;3(12):e254. doi: 10.1371/journal.pcbi.0030254. Epub 2007 Nov 14. PLoS Comput Biol. 2007. PMID: 18166073 Free PMC article. - Selective constraint, background selection, and mutation accumulation variability within and between human populations.
Hodgkinson A, Casals F, Idaghdour Y, Grenier JC, Hernandez RD, Awadalla P. Hodgkinson A, et al. BMC Genomics. 2013 Jul 23;14:495. doi: 10.1186/1471-2164-14-495. BMC Genomics. 2013. PMID: 23875710 Free PMC article. - Natural Selection and Functional Potentials of Human Noncoding Elements Revealed by Analysis of Next Generation Sequencing Data.
Jha P, Lu D, Xu S. Jha P, et al. PLoS One. 2015 Jun 8;10(6):e0129023. doi: 10.1371/journal.pone.0129023. eCollection 2015. PLoS One. 2015. PMID: 26053627 Free PMC article. - The promise of comparative genomics in mammals.
O'Brien SJ, Menotti-Raymond M, Murphy WJ, Nash WG, Wienberg J, Stanyon R, Copeland NG, Jenkins NA, Womack JE, Marshall Graves JA. O'Brien SJ, et al. Science. 1999 Oct 15;286(5439):458-62, 479-81. doi: 10.1126/science.286.5439.458. Science. 1999. PMID: 10521336 Review. - Cis-acting regulatory variation in the human genome.
Pastinen T, Hudson TJ. Pastinen T, et al. Science. 2004 Oct 22;306(5696):647-50. doi: 10.1126/science.1101659. Science. 2004. PMID: 15499010 Review.
Cited by
- IGF1R variants associated with isolated single suture craniosynostosis.
Cunningham ML, Horst JA, Rieder MJ, Hing AV, Stanaway IB, Park SS, Samudrala R, Speltz ML. Cunningham ML, et al. Am J Med Genet A. 2011 Jan;155A(1):91-7. doi: 10.1002/ajmg.a.33781. Am J Med Genet A. 2011. PMID: 21204214 Free PMC article. - Gene family information facilitates variant interpretation and identification of disease-associated genes in neurodevelopmental disorders.
Lal D, May P, Perez-Palma E, Samocha KE, Kosmicki JA, Robinson EB, Møller RS, Krause R, Nürnberg P, Weckhuysen S, De Jonghe P, Guerrini R, Niestroj LM, Du J, Marini C; EuroEPINOMICS-RES Consortium; Ware JS, Kurki M, Gormley P, Tang S, Wu S, Biskup S, Poduri A, Neubauer BA, Koeleman BPC, Helbig KL, Weber YG, Helbig I, Majithia AR, Palotie A, Daly MJ. Lal D, et al. Genome Med. 2020 Mar 17;12(1):28. doi: 10.1186/s13073-020-00725-6. Genome Med. 2020. PMID: 32183904 Free PMC article. - Relaxed Selection During a Recent Human Expansion.
Peischl S, Dupanloup I, Foucal A, Jomphe M, Bruat V, Grenier JC, Gouy A, Gilbert KJ, Gbeha E, Bosshard L, Hip-Ki E, Agbessi M, Hodgkinson A, Vézina H, Awadalla P, Excoffier L. Peischl S, et al. Genetics. 2018 Feb;208(2):763-777. doi: 10.1534/genetics.117.300551. Epub 2017 Nov 29. Genetics. 2018. PMID: 29187508 Free PMC article. - Lysoplex: An efficient toolkit to detect DNA sequence variations in the autophagy-lysosomal pathway.
Di Fruscio G, Schulz A, De Cegli R, Savarese M, Mutarelli M, Parenti G, Banfi S, Braulke T, Nigro V, Ballabio A. Di Fruscio G, et al. Autophagy. 2015;11(6):928-38. doi: 10.1080/15548627.2015.1043077. Autophagy. 2015. PMID: 26075876 Free PMC article. - Whole-exome sequencing reveals a rapid change in the frequency of rare functional variants in a founding population of humans.
Casals F, Hodgkinson A, Hussin J, Idaghdour Y, Bruat V, de Maillard T, Grenier JC, Gbeha E, Hamdan FF, Girard S, Spinella JF, Larivière M, Saillour V, Healy J, Fernández I, Sinnett D, Michaud JL, Rouleau GA, Haddad E, Le Deist F, Awadalla P. Casals F, et al. PLoS Genet. 2013;9(9):e1003815. doi: 10.1371/journal.pgen.1003815. Epub 2013 Sep 26. PLoS Genet. 2013. PMID: 24086152 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources