Prospective comparison of clinical and genomic multivariate predictors of response to neoadjuvant chemotherapy in breast cancer - PubMed (original) (raw)
Comparative Study
Prospective comparison of clinical and genomic multivariate predictors of response to neoadjuvant chemotherapy in breast cancer
Jae K Lee et al. Clin Cancer Res. 2010.
Abstract
Purpose: Several different multivariate prediction models using routine clinical variables or multigene signatures have been proposed to predict pathologic complete response to combination chemotherapy in breast cancer. Our goal was to compare the performance of four conceptually different predictors in an independent cohort of patients.
Experimental design: Gene expression profiling was done on fine-needle aspirations of 100 stage I to III breast cancers before preoperative paclitaxel, 5-fluorouracil, doxorubicin, and cyclophosphamide combination chemotherapy. Pathologic response was correlated with prediction results from a clinical nomogram, a human cancer-derived genomic predictor (DLDA30), a cell line-based genomic predictor [in vitro coexpression extrapolation (COXEN)], and an optimized cell line-derived (in vivo COXEN) predictor. None of the 100 test cases were used in the development of these predictors.
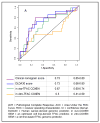
Results: The in vitro COXEN using a combination of four individual drug sensitivity predictions derived from cell lines was not predictive [area under the receiver operator characteristic curve (AUC), 0.5; 95% confidence interval, (95% CI), 0.41-0.59]. The clinical nomogram (AUC, 0.73; 95% CI, 0.65-0.80) and the DLDA30 (AUC, 0.73; 95% CI, 0.66-0.80) genomic predictor had similar performances. The in vivo COXEN that used informative genes from cell lines but was trained on a separate human data set also showed significant predictive value (AUC, 0.67; 95% CI, 0.60-0.74). These three different prediction scores correlated with each other and were significant in univariate but not in multivariate analysis.
Conclusions: Three conceptually different predictors performed similarly in this validation study and tended to identify the same patients as responders. A genomic predictor that relied solely on a composite of individual drug sensitivity predictions from cell lines did not show any predictive value.
Conflict of interest statement
Conflicts of interest
The authors declared no conflicts of interest.
Figures
Figure 1. Receiver Operating Characteristics (ROC) curve analysis of the four conceptually different chemotherapy response predictors
ROC curves are shown for the clinical nomogram, the DLDA30 predictor, the in vitro combined T/FAC COXEN GEM and the in vivo combined T/FAC COXEN GEM.
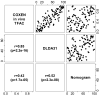
Figure 2. Correlation between ranks of prediction scores
Ranks of predicted scores of the in vivo-GEM, DLDA30 predictor and clinical nomogram were plotted against each other. Rank-based Spearman correlation and p-value were calculated for each pair of comparisons.
Similar articles
- Evaluation of a 30-gene paclitaxel, fluorouracil, doxorubicin, and cyclophosphamide chemotherapy response predictor in a multicenter randomized trial in breast cancer.
Tabchy A, Valero V, Vidaurre T, Lluch A, Gomez H, Martin M, Qi Y, Barajas-Figueroa LJ, Souchon E, Coutant C, Doimi FD, Ibrahim NK, Gong Y, Hortobagyi GN, Hess KR, Symmans WF, Pusztai L. Tabchy A, et al. Clin Cancer Res. 2010 Nov 1;16(21):5351-61. doi: 10.1158/1078-0432.CCR-10-1265. Epub 2010 Sep 9. Clin Cancer Res. 2010. PMID: 20829329 Free PMC article. Clinical Trial. - Pharmacogenomic predictor of sensitivity to preoperative chemotherapy with paclitaxel and fluorouracil, doxorubicin, and cyclophosphamide in breast cancer.
Hess KR, Anderson K, Symmans WF, Valero V, Ibrahim N, Mejia JA, Booser D, Theriault RL, Buzdar AU, Dempsey PJ, Rouzier R, Sneige N, Ross JS, Vidaurre T, Gómez HL, Hortobagyi GN, Pusztai L. Hess KR, et al. J Clin Oncol. 2006 Sep 10;24(26):4236-44. doi: 10.1200/JCO.2006.05.6861. Epub 2006 Aug 8. J Clin Oncol. 2006. PMID: 16896004 - Clinical evaluation of chemotherapy response predictors developed from breast cancer cell lines.
Liedtke C, Wang J, Tordai A, Symmans WF, Hortobagyi GN, Kiesel L, Hess K, Baggerly KA, Coombes KR, Pusztai L. Liedtke C, et al. Breast Cancer Res Treat. 2010 Jun;121(2):301-9. doi: 10.1007/s10549-009-0445-7. Epub 2009 Jul 15. Breast Cancer Res Treat. 2010. PMID: 19603265 - Gene expression profiles predict complete pathologic response to neoadjuvant paclitaxel and fluorouracil, doxorubicin, and cyclophosphamide chemotherapy in breast cancer.
Ayers M, Symmans WF, Stec J, Damokosh AI, Clark E, Hess K, Lecocke M, Metivier J, Booser D, Ibrahim N, Valero V, Royce M, Arun B, Whitman G, Ross J, Sneige N, Hortobagyi GN, Pusztai L. Ayers M, et al. J Clin Oncol. 2004 Jun 15;22(12):2284-93. doi: 10.1200/JCO.2004.05.166. Epub 2004 May 10. J Clin Oncol. 2004. PMID: 15136595 - Is there any role for new prognostic markers in breast cancer?
Barnadas A. Barnadas A. Clin Transl Oncol. 2012 Mar;14(3):161-2. doi: 10.1007/s12094-012-0778-2. Clin Transl Oncol. 2012. PMID: 22374417 Review. No abstract available.
Cited by
- A systematic evaluation of multi-gene predictors for the pathological response of breast cancer patients to chemotherapy.
Shen K, Song N, Kim Y, Tian C, Rice SD, Gabrin MJ, Symmans WF, Pusztai L, Lee JK. Shen K, et al. PLoS One. 2012;7(11):e49529. doi: 10.1371/journal.pone.0049529. Epub 2012 Nov 21. PLoS One. 2012. PMID: 23185353 Free PMC article. - High-throughput molecular analysis from leftover of fine needle aspiration cytology of mammographically detected breast cancer.
Annaratone L, Marchiò C, Renzulli T, Castellano I, Cantarella D, Isella C, Macrì L, Mariscotti G, Balmativola D, Cantanna E, Deambrogio C, Pietribiasi F, Arisio R, Schmitt F, Medico E, Sapino A. Annaratone L, et al. Transl Oncol. 2012 Jun;5(3):180-9. doi: 10.1593/tlo.11343. Epub 2012 Jun 1. Transl Oncol. 2012. PMID: 22741037 Free PMC article. - Importance of pre-analytical steps for transcriptome and RT-qPCR analyses in the context of the phase II randomised multicentre trial REMAGUS02 of neoadjuvant chemotherapy in breast cancer patients.
de Cremoux P, Valet F, Gentien D, Lehmann-Che J, Scott V, Tran-Perennou C, Barbaroux C, Servant N, Vacher S, Sigal-Zafrani B, Mathieu MC, Bertheau P, Guinebretière JM, Asselain B, Marty M, Spyratos F. de Cremoux P, et al. BMC Cancer. 2011 Jun 1;11:215. doi: 10.1186/1471-2407-11-215. BMC Cancer. 2011. PMID: 21631949 Free PMC article. Clinical Trial. - A prognostic 4-gene expression signature for patients with HER2-negative breast cancer receiving taxane and anthracycline-based chemotherapy.
Cheng P, Wang Z, Hu G, Huang Q, Han M, Huang J. Cheng P, et al. Oncotarget. 2017 Oct 17;8(61):103327-103339. doi: 10.18632/oncotarget.21872. eCollection 2017 Nov 28. Oncotarget. 2017. PMID: 29262565 Free PMC article.
References
- Potti A, Dressman HK, Bild A, et al. Genomic signatures to guide the use of chemotherapeutics. Nat Med. 2006;12:1294–300. - PubMed
- Bonnefoi H, Potti A, Delorenzi M, et al. Validation of gene signatures that predict the response of breast cancer to neoadjuvant chemotherapy: a substudy of the EORTC 10994/BIG 00-01 clinical trial. Lancet Oncol. 2007;8:1071–8. - PubMed
- Coombes KR, Wang J, Baggerly KA. Microarrays: retracing steps. Nat Med. 2007;13:1276–7. author reply 7-8. - PubMed
- Hsu DS, Balakumaran BS, Acharya CR, et al. Pharmacogenomic strategies provide a rational approach to the treatment of cisplatin-resistant patients with advanced cancer. J Clin Oncol. 2007;25:4350–7. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA106290-03/CA/NCI NIH HHS/United States
- R01 CA075115/CA/NCI NIH HHS/United States
- R01 HL081690/HL/NHLBI NIH HHS/United States
- R01 HL081690-04/HL/NHLBI NIH HHS/United States
- R01CA075115/CA/NCI NIH HHS/United States
- R29 CA075115/CA/NCI NIH HHS/United States
- R29 CA075115-05/CA/NCI NIH HHS/United States
- R01HL081690/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical