DNA methylation-mediated down-regulation of DNA methyltransferase-1 (DNMT1) is coincident with, but not essential for, global hypomethylation in human placenta - PubMed (original) (raw)
. 2010 Mar 26;285(13):9583-9593.
doi: 10.1074/jbc.M109.064956. Epub 2010 Jan 13.
Nick C Wong 2, Mandy Sibson 2, Hong-Kiat Ng 2, Ruth Morley 2, Ursula Manuelpillai 3, Thomas Down 4, Vardhman K Rakyan 5, Stephan Beck 6, Stefan Hiendleder 7, Claire T Roberts 8, Jeffrey M Craig 1, Richard Saffery 9
Affiliations
- PMID: 20071334
- PMCID: PMC2843208
- DOI: 10.1074/jbc.M109.064956
DNA methylation-mediated down-regulation of DNA methyltransferase-1 (DNMT1) is coincident with, but not essential for, global hypomethylation in human placenta
Boris Novakovic et al. J Biol Chem. 2010.
Abstract
The genome of extraembryonic tissue, such as the placenta, is hypomethylated relative to that in somatic tissues. However, the origin and role of this hypomethylation remains unclear. The DNA methyltransferases DNMT1, -3A, and -3B are the primary mediators of the establishment and maintenance of DNA methylation in mammals. In this study, we investigated promoter methylation-mediated epigenetic down-regulation of DNMT genes as a potential regulator of global methylation levels in placental tissue. Although DNMT3A and -3B promoters lack methylation in all somatic and extraembryonic tissues tested, we found specific hypermethylation of the maintenance DNA methyltransferase (DNMT1) gene and found hypomethylation of the DNMT3L gene in full term and first trimester placental tissues. Bisulfite DNA sequencing revealed monoallelic methylation of DNMT1, with no evidence of imprinting (parent of origin effect). In vitro reporter experiments confirmed that DNMT1 promoter methylation attenuates transcriptional activity in trophoblast cells. However, global hypomethylation in the absence of DNMT1 down-regulation is apparent in non-primate placentas and in vitro derived human cytotrophoblast stem cells, suggesting that DNMT1 down-regulation is not an absolute requirement for genomic hypomethylation in all instances. These data represent the first demonstration of methylation-mediated regulation of the DNMT1 gene in any system and demonstrate that the unique epigenome of the human placenta includes down-regulation of DNMT1 with concomitant hypomethylation of the DNMT3L gene. This strongly implicates epigenetic regulation of the DNMT gene family in the establishment of the unique epigenetic profile of extraembryonic tissue in humans.
Figures
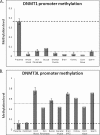
FIGURE 1.
SEQUENOM MassARRAY EpiTYPER analysis of DNMT1 and DNMT3L genes in human placenta and somatic tissues. A, placenta-specific methylation of the DNMT1 promoter assessed using the DNMT1_5 assay, spanning 230 bp from −133 to +97 relative to the transcription start site. Mean methylation in somatic tissues was 4.4% (across eight different tissues), whereas full term placental tissue shows a mean methylation level of 36.6% (n = 8 placentas). B, placenta-specific hypomethylation of the DNMT3L gene assessed using the DNMT3L_1 assay spanning 371 bp, −18/+353 relative to the transcription start site. Mean methylation in term placenta and purified cytotrophoblasts was 14 and 13%, respectively, whereas the mean for other tissues was 53% (n = 8) across three analyzable CpG units.
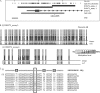
FIGURE 2.
Monoallelic methylation of the human DNMT1 gene. A, location of the DNMT1 CpG island/promoter DNA methylation assays used for bisulfite sequencing. Genomic coordinates on chromosome 19 are shown, along with location of individual CpG dinucleotides (dashes) and CpG island location (gray bar) in relation to the DNMT 1 gene (exon 1 shown as a blue box, intron shown as an arrowed line) according to the University of California Santa Cruz genome browser (hg_18 assembly). The arrows denote transcriptional direction. The location of rs8112895 SNP in the DNMT1_assay_4 is also shown. B, representative DNA methylation of a single full term human placental sample for assay_1 (i) and assay_4 (ii) showing a monoallelic pattern of methylation within the human placenta. Between eight and 12 individual clones were sequenced for each sample. The circles correspond to CpG sites denoted by black dashes in A. Closed circles, methylation; open circles, lack of methylation. Mean methylation levels seen at each CpG site in multiple placental samples for each assay are shown as shaded bars between 0 and 100% (n = 4). Missing circles indicate CpG sites for which no information was obtained. C, DNMT1_4 methylation assay spanning SNP rs8112895 was used for bisulfite sequencing methylation analysis in purified first trimester cytotrophoblast cells. i, reference genomic DNA sequence surrounding rs8112895 SNP (R) and CpG dinucleotides (boxed). ii, individual clone sequences showing the presence of bisulfite-converted CG sites (TG) and unconverted CG sites (CG) in both maternal and paternal alleles from this heterozygous sample. This confirms a lack of parent of origin of the observed DNMT1 promoter methylation.
FIGURE 3.
Distribution of monoallelic methylation of the DNMT1 promoter in first trimester placental tissue and trophoblasts. Bisulfite sequencing data obtained using the DNMT1_1 assay in the following tissues: purified cytotrophoblasts (A) and uncultured (B) and cultured (C) (n = 10 passages) CVS biopsies. i, examples of data from individual samples; ii, mean methylation level for biological replicates (cytotrophoblasts, n = 2; uncultured and cultured CVS, n = 6). Shown are both sperm (D) and whole blood (E) lack of DNMT1 methylation as assessed by this methodology.
FIGURE 4.
DNMT1 promoter methylation is restricted to primates. Bisulfite DNA sequencing confirmed promoter methylation of the DNMT1 gene in full term baboon (A) and marmoset (B) placental tissue, with no evidence for methylation in mouse (C), bovine, or guinea pig placental tissue (data not shown).
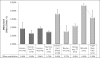
FIGURE 5.
Widely variable DNMT1 expression in placental tissue and derived cells. A, DNMT1 expression levels in methylated samples (orange bars) versus non-methylated (yellow bars) relative to the mean full term placenta DNMT1 expression level. Data are shown for full term placenta (Plac; n = 4), uncultured first trimester CVS tissue (uCVS; n = 2), purified first trimester cytotrophoblasts (CT; n = 2), choriocarcinoma cell lines (CCA; n = 3), SV40-immortalized trophoblast cell line HIPEC65, and placental fibroblasts (pFIB; n = 1). No obvious correlation between gene expression and promoter methylation was apparent. B, decreasing DNMT1 expression was seen in following culturing of CVS tissue for 10 passages (cCVS; n = 2) and with syncitialization of BeWo cell lines (BeWo-post; n = 2). Expression level is relative to the mean of uncultured CVS tissue. The colored bars denote methylation status as per A. C, correlation between DNMT1 expression level and methylation status in CVS tissue (pre- and post-culturing). Culturing for 10 passages results in a decrease in DNMT1 expression, whereas promoter methylation is increased. The correlation co-efficient is −0.797, suggesting an inverse relationship between methylation and expression levels in this system.
FIGURE 6.
Methylation of the DNMT1 promoter directly attenuates gene transcriptional level. A, position of the cloned DNMT1 promoter region (pale blue bar; −536/+374 relative to the transcription start site). Shown are genome coordinates, GC percentage, location of individual CpG dinucleotides, CpG island (green bar), and DNMT1 gene (exons shown as blue boxes, intron as an arrowed line) according to the University of California Santa Cruz genome browser (hg_18 assembly). The arrows denote transcriptional direction. B, luciferase reporter constructs were transfected into human JAR cells with or without prior in vitro methylation with SssI, HpaII, or HhaI methylases (n = 8 for each). A 15-fold increase in promoter activity was detected for the unmethylated promoter region relative to the promoterless vector. This activity was abolished with in vitro methylation prior to transfection (SssI, HpaII, and HhaI methylases). Luminescence values were normalized to Renilla luciferase to correct for transfection efficiency, and all data are displayed relative to the mean basal promoter activity of the pGL3:basic vector. The error bars denote 95% confidence intervals. Statistical analysis for comparing between transfected groups was carried out using a two-tailed Student's t test with unequal variance.
FIGURE 7.
Methylation status of DNMT1 does not predict global 5MeC levels in extraembryonic tissue. Global 5MeC content was measured in a variety of placental and umbilical cord blood cells. Human full term (FT), first trimester, full term pre-eclamptic (PE), and full term marmoset tissue (with DNMT1 promoter methylation) show mean global 5MeC levels of <3% (range 2.64–2.93%). Despite lacking methylation of the equivalent DNMT1 gene promoter, bovine and guinea pig FT placental tissues show a similar mean global 5MeC level (2.77 and 3.1% methylation, respectively), indicating that DNMT1 promoter methylation is not an absolute prerequisite for reduced genomic methylation in extraembryonic tissue. Human cord blood (also lacking DNMT1 promoter methylation) shows a mean global 5MeC content of 3.82%, consistent with previous reports in peripheral blood (5). Human embryonic stem cells showed the highest global 5MeC level of all tissues/cells examined with a mean of 4.33%. Differentiation of these cells into cytotrophoblast stem cells was associated with a reduction in global methylation levels (mean 3.59%) in the absence of concomitant DNMT1 promoter methylation (data not shown). However, the small sample numbers resulted in a lack of significance when tested using a paired Student's t test. The dark bars correspond to samples with DNMT1 promoter methylation, whereas light bars identify samples lacking DNMT1 promoter methylation.
Similar articles
- Regulation of lineage specific DNA hypomethylation in mouse trophectoderm.
Oda M, Oxley D, Dean W, Reik W. Oda M, et al. PLoS One. 2013 Jun 25;8(6):e68846. doi: 10.1371/journal.pone.0068846. Print 2013. PLoS One. 2013. PMID: 23825703 Free PMC article. - Distinct roles of DNMT1-dependent and DNMT1-independent methylation patterns in the genome of mouse embryonic stem cells.
Li Z, Dai H, Martos SN, Xu B, Gao Y, Li T, Zhu G, Schones DE, Wang Z. Li Z, et al. Genome Biol. 2015 Jun 2;16(1):115. doi: 10.1186/s13059-015-0685-2. Genome Biol. 2015. PMID: 26032981 Free PMC article. - Decreased expression and altered methylation of syncytin-1 gene in human placentas associated with preeclampsia.
Zhuang XW, Li J, Brost BC, Xia XY, Chen HB, Wang CX, Jiang SW. Zhuang XW, et al. Curr Pharm Des. 2014;20(11):1796-802. doi: 10.2174/13816128113199990541. Curr Pharm Des. 2014. PMID: 23888950 - The Roles of Human DNA Methyltransferases and Their Isoforms in Shaping the Epigenome.
Gujar H, Weisenberger DJ, Liang G. Gujar H, et al. Genes (Basel). 2019 Feb 23;10(2):172. doi: 10.3390/genes10020172. Genes (Basel). 2019. PMID: 30813436 Free PMC article. Review. - Specific or not specific recruitment of DNMTs for DNA methylation, an epigenetic dilemma.
Hervouet E, Peixoto P, Delage-Mourroux R, Boyer-Guittaut M, Cartron PF. Hervouet E, et al. Clin Epigenetics. 2018 Feb 9;10:17. doi: 10.1186/s13148-018-0450-y. eCollection 2018. Clin Epigenetics. 2018. PMID: 29449903 Free PMC article. Review.
Cited by
- Reduced syncytin-1 expression levels in placental syndromes correlates with epigenetic hypermethylation of the ERVW-1 promoter region.
Ruebner M, Strissel PL, Ekici AB, Stiegler E, Dammer U, Goecke TW, Faschingbauer F, Fahlbusch FB, Beckmann MW, Strick R. Ruebner M, et al. PLoS One. 2013;8(2):e56145. doi: 10.1371/journal.pone.0056145. Epub 2013 Feb 14. PLoS One. 2013. PMID: 23457515 Free PMC article. - The Placental Function Beyond Pregnancy: Insights from Latin America.
Carrasco-Wong I, González-Ortiz M, Araujo GG, Lima VV, Giachini FR, Stojanova J, Moller A, Martín SS, Escudero P, Damiano AE, Sosa-Macias M, Galaviz-Hernandez C, Teran E, Escudero C; on behalf RIVATREM. Carrasco-Wong I, et al. Adv Exp Med Biol. 2023;1428:287-307. doi: 10.1007/978-3-031-32554-0_13. Adv Exp Med Biol. 2023. PMID: 37466779 Review. - Maternal PUFAs, Placental Epigenetics, and Their Relevance to Fetal Growth and Brain Development.
Basak S, Duttaroy AK. Basak S, et al. Reprod Sci. 2023 Feb;30(2):408-427. doi: 10.1007/s43032-022-00989-w. Epub 2022 Jun 8. Reprod Sci. 2023. PMID: 35676498 Review. - Supplemental Herbal Choline Increases 5-hmC DNA on Whole Blood from Pregnant Ewes and Offspring.
Roque-Jiménez JA, Mendoza-Martínez GD, Vázquez-Valladolid A, Guerrero-González ML, Flores-Ramírez R, Pinos-Rodriguez JM, Loor JJ, Relling AE, Lee-Rangel HA. Roque-Jiménez JA, et al. Animals (Basel). 2020 Jul 27;10(8):1277. doi: 10.3390/ani10081277. Animals (Basel). 2020. PMID: 32727060 Free PMC article.
References
- Bestor T. H. (2000) Hum. Mol. Genet. 9, 2395–2402 - PubMed
- Aapola U., Kawasaki K., Scott H. S., Ollila J., Vihinen M., Heino M., Shintani A., Kawasaki K., Minoshima S., Krohn K., Antonarakis S. E., Shimizu N., Kudoh J., Peterson P. (2000) Genomics 65, 293–298 - PubMed
- Suetake I., Shinozaki F., Miyagawa J., Takeshima H., Tajima S. (2004) J. Biol. Chem. 279, 27816–27823 - PubMed
- Fuke C., Shimabukuro M., Petronis A., Sugimoto J., Oda T., Miura K., Miyazaki T., Ogura C., Okazaki Y., Jinno Y. (2004) Ann. Hum. Genet. 68, 196–204 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources