Heterologous quaternary structure of CXCL12 and its relationship to the CC chemokine family - PubMed (original) (raw)
Heterologous quaternary structure of CXCL12 and its relationship to the CC chemokine family
James W Murphy et al. Proteins. 2010 Apr.
No abstract available
Figures
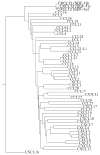
Figure 1
Phylogenetic tree calculated using the neighbor-joining (NJ) method representing all human chemokines and their isoforms. The tree is based on the multiple sequence alignment. The three isoforms of CXCL12 (SDF-1α, β, and γ) are indicated at the uppermost portion of the figure.
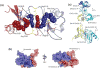
Figure 2
Crystal structures of CXCL12. (a) Cartoon representation of the single protein chain of the asymmetric unit in the P3221 space group CXCL12 crystals. Chemokine structural elements are labeled. (b) The protein composition of the asymmetric unit in the P212121 space group CXCL12 crystals. Ten monomers are arranged as an elongated decamer and form five CC-like dimers. (c) The semi-transparent surface representation of the asymmetric unit of the P3221 space group of CXCL12 and a symmetry mate forms a CXC dimer that interact through their α-helices and β-strand-1. (d) Semi-transparent surface of two representative chains in the decamer of CXCL12 which form a CC chemokine-like dimer and interact with their amino termini and 30’s loops. (e) CXC dimer of CXCL12 (upper panel) compared to CC dimer of CXCL12 (lower panel). Both dimers were aligned using only Chain A of each in the alignment algorithm.
Figure 3
Intersubunit contacts of CXCL12 dimers. (a) Arginine zipper motif forms the interactions in the amino terminal regions of the CC-like CXCL12 dimer. Cysteines are colored yellow, arginines from chains A and B are colored purple and red, respectively. (b) Crystallographic packing between CC-dimers. Cartoon representation with semi-transparent surface of two representative chains in the decamer of CXCL12 in between each CC chemokine-like dimer. (c) Overlay of the CC-like dimer of CXCL12 and the CC dimer of CCL5. Chain A from each dimer was aligned. Glutamic acid 15 from CXCL12 and isoleucine 15 from CCL5 are shown as sticks to indicate the 180° rotation of the carbon backbone.
Similar articles
- Structural biology. Crystal structure of the chemokine receptor CXCR4 in complex with a viral chemokine.
Qin L, Kufareva I, Holden LG, Wang C, Zheng Y, Zhao C, Fenalti G, Wu H, Han GW, Cherezov V, Abagyan R, Stevens RC, Handel TM. Qin L, et al. Science. 2015 Mar 6;347(6226):1117-22. doi: 10.1126/science.1261064. Epub 2015 Jan 22. Science. 2015. PMID: 25612609 Free PMC article. - Structures of human CCL18, CCL3, and CCL4 reveal molecular determinants for quaternary structures and sensitivity to insulin-degrading enzyme.
Liang WG, Ren M, Zhao F, Tang WJ. Liang WG, et al. J Mol Biol. 2015 Mar 27;427(6 Pt B):1345-1358. doi: 10.1016/j.jmb.2015.01.012. Epub 2015 Jan 28. J Mol Biol. 2015. PMID: 25636406 Free PMC article. - Investigation of CC and CXC chemokine quaternary state mutants.
Jin H, Hayes GL, Darbha NS, Meyer E, LiWang PJ. Jin H, et al. Biochem Biophys Res Commun. 2005 Dec 16;338(2):987-99. doi: 10.1016/j.bbrc.2005.10.062. Epub 2005 Oct 21. Biochem Biophys Res Commun. 2005. PMID: 16256937 - Evolution of CC chemokines in teleost fish: a case study in gene duplication and implications for immune diversity.
Peatman E, Liu Z. Peatman E, et al. Immunogenetics. 2007 Aug;59(8):613-23. doi: 10.1007/s00251-007-0228-4. Epub 2007 May 31. Immunogenetics. 2007. PMID: 17541578 Review. - The relationship between oligomeric state and protein function.
Griffin MD, Gerrard JA. Griffin MD, et al. Adv Exp Med Biol. 2012;747:74-90. doi: 10.1007/978-1-4614-3229-6_5. Adv Exp Med Biol. 2012. PMID: 22949112 Review.
Cited by
- Structural insights into CXCR4 modulation and oligomerization.
Saotome K, McGoldrick LL, Ho JH, Ramlall TF, Shah S, Moore MJ, Kim JH, Leidich R, Olson WC, Franklin MC. Saotome K, et al. Nat Struct Mol Biol. 2024 Sep 23. doi: 10.1038/s41594-024-01397-1. Online ahead of print. Nat Struct Mol Biol. 2024. PMID: 39313635 - Structural insight into the evolution of a new chemokine family from zebrafish.
Rajasekaran D, Fan C, Meng W, Pflugrath JW, Lolis EJ. Rajasekaran D, et al. Proteins. 2014 May;82(5):708-16. doi: 10.1002/prot.24380. Epub 2013 Dec 14. Proteins. 2014. PMID: 23900850 Free PMC article. - Interaction and dynamics of chemokine receptor CXCR4 binding with CXCL12 and hBD-3.
Penfield J, Zhang L. Penfield J, et al. Commun Chem. 2024 Sep 13;7(1):205. doi: 10.1038/s42004-024-01280-6. Commun Chem. 2024. PMID: 39271963 Free PMC article. - Structural basis for chemokine recognition by a G protein-coupled receptor and implications for receptor activation.
Ziarek JJ, Kleist AB, London N, Raveh B, Montpas N, Bonneterre J, St-Onge G, DiCosmo-Ponticello CJ, Koplinski CA, Roy I, Stephens B, Thelen S, Veldkamp CT, Coffman FD, Cohen MC, Dwinell MB, Thelen M, Peterson FC, Heveker N, Volkman BF. Ziarek JJ, et al. Sci Signal. 2017 Mar 21;10(471):eaah5756. doi: 10.1126/scisignal.aah5756. Sci Signal. 2017. PMID: 28325822 Free PMC article. - Structural perspectives on antimicrobial chemokines.
Nguyen LT, Vogel HJ. Nguyen LT, et al. Front Immunol. 2012 Dec 28;3:384. doi: 10.3389/fimmu.2012.00384. eCollection 2012. Front Immunol. 2012. PMID: 23293636 Free PMC article.
References
- Rot A, von Andrian UH. Chemokines in innate and adaptive host defense: basic chemokinese grammar for immune cells. Annu Rev Immunol. 2004;22:891–928. - PubMed
- Viola A, Luster AD. Chemokines and Their Receptors: Drug Targets in Immunity and Inflammation. Annual Review of Pharmacology and Toxicology. 2008;48(1):171–197. - PubMed
- Fernandez EJ, Wilken J, Thompson DA, Peiper SC, Lolis E. Comparison of the structure of vMIP-II with eotaxin-1, RANTES, and MCP-3 suggests a unique mechanism for CCR3 activation. Biochemistry. 2000;39(42):12837–12844. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI082295-03/AI/NIAID NIH HHS/United States
- AI082295/AI/NIAID NIH HHS/United States
- P30 EB009998/EB/NIBIB NIH HHS/United States
- R01 AI082295-01/AI/NIAID NIH HHS/United States
- R01 AI082295/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases