Mammalian sirtuins: biological insights and disease relevance - PubMed (original) (raw)
Review
Mammalian sirtuins: biological insights and disease relevance
Marcia C Haigis et al. Annu Rev Pathol. 2010.
Abstract
Aging is accompanied by a decline in the healthy function of multiple organ systems, leading to increased incidence and mortality from diseases such as type II diabetes mellitus, neurodegenerative diseases, cancer, and cardiovascular disease. Historically, researchers have focused on investigating individual pathways in isolated organs as a strategy to identify the root cause of a disease, with hopes of designing better drugs. Studies of aging in yeast led to the discovery of a family of conserved enzymes known as the sirtuins, which affect multiple pathways that increase the life span and the overall health of organisms. Since the discovery of the first known mammalian sirtuin, SIRT1, 10 years ago, there have been major advances in our understanding of the enzymology of sirtuins, their regulation, and their ability to broadly improve mammalian physiology and health span. This review summarizes and discusses the advances of the past decade and the challenges that will confront the field in the coming years.
Figures
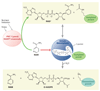
Figure 1
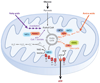
The sirtuin deacetylation reaction and regulation by stress and nutrition. Unlike type I and II deacetylases, which hydrolyze the acetyl group on a substrate, sirtuin deacetylases catalyze an unprecedented two-step biological reaction that consumes nicotinamide adenine dinucleotide (NAD+) and releases nicotinamide (NAM), _O_-acetyl-ADP-ribose (AADPR), and the deacetylated substrate. Amide-to-ester acyltransfer is unfavorable, but hydrolysis of NAD+ can provide a favorable driving force for the overall sirtuin reaction. Evidence favors a mechanism in which electrophilic capture of the acetyl oxygen in an ADP-ribosyltransfer reaction forms an ADP-ribose peptide-imidate complex. This intermediate may last for a few seconds, enough time for NAM to enter the C-pocket and catalyze a reverse reaction. Activation of sirtuins can be facilitated by the removal of NAM and its conversion to NAD+ by PNC1 (yeast and simple metazoans) or NAMPT (mammals and alpha proteobacteria), two genes upregulated by stress and nutrient limitation.
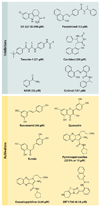
Figure 2
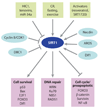
Chemical inhibitors and activators of SIRT1. Over the past 10 years, a variety of small-molecule SIRT1-activating compounds (STACs) or inhibitors have been published. The known IC50s and EC1.5s for SIRT1 are shown in parentheses. Of all the inhibitors, only nicotinamide (NAM) is a physiological inhibitor, although analogs of NAM can activate sirtuins, apparently by occluding the C-pocket (see Figure 1). Polyphenolic activators such as resveratrol appear to bind the same site and activate via the same mechanism (i.e., primarily a lowering of the Michaelis constant effect) as that used by more potent activators such as SRT1720. The potency of inhibition is expressed as IC50 (the concentration to inhibit 50% activity), and activation is expressed as EC1.5 (the concentration to activate 1.5-fold).
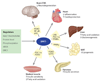
Figure 3
SIRT1 regulation of age-related physiology. SIRT1 activity can be regulated through NAD+ (nicotinamide adenine dinucleotide) and nicotinamide (NAM) concentrations, by SIRT1 protein level, and by phosphorylation; SIRT1 can be activated by active regulator of SIRT1 (AROS) and inhibited by DBC1 (deleted in breast cancer 1). SIRT1 activation promotes survival of neurons and protects cardiomyocytes from death. In the liver, SIRT1 promotes fatty acid oxidation and gluconeogenesis during nutrient deprivation via LXR, PGC-1α, and PPARα. In white adipose tissue (WAT), SIRT1 decreases fat storage by repressing PPARγ. SIRT1 promotes insulin secretion and pancreatic beta cell survival by suppressing UCP2 and interacting with FOXO, respectively. In skeletal muscle, SIRT1 promotes mitochondrial biogenesis through the activation of PGC-1α. Abbreviations: CNS, central nervous system; FOXO, forkhead box transcription factor, subgroup O; LXR, liver X receptor; NF-κB, nuclear factor kappa B; PGC-1α, peroxisome proliferator-activated receptor gamma coactivator 1 alpha; PPARα, peroxisome proliferators-activated receptor alpha; UCP2, uncoupling protein 2. Adapted from Reference .
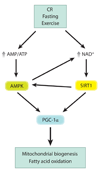
Figure 4
The SIRT1-AMPK metabolic control network. Conditions of perceived energy deprivation, such as fasting, calorie restriction (CR), and exercise, increase the AMP/ATP ratio and activate AMP-activated protein kinase (AMPK). Energy deprivation also increases nicotinamide adenine dinucleotide (NAD+) levels and activates the NAD+-dependent deacetylase activity of SIRT1. Activated AMPK and SIRT1 converge by activating peroxisome proliferator-activated receptor gamma coactivator 1 alpha (PGC-1α) via phosphorylation and deacetylation, respectively, to induce mitochondrial biogenesis and fatty acid oxidation. Cross talk in this pathway occurs because AMPK activity increases NAD+, and SIRT1 also activates AMPK.
Figure 5
Network of the mitochondrial sirtuins (SIRT3–5). Mitochondria can metabolize fuels, such as fatty acids, amino acids, and pyruvate, derived from glucose. Electrons pass through electron transport complexes (I–IV), generating a proton gradient that is used to drive ATP synthase to generate ATP. SIRT3 binds to complex I, regulating its activity and energy levels in the cell. SIRT3 also binds and deacetylates acetyl-CoA synthetase 2 (AceCS2) and glutamate dehydrogenase (GDH), activating their enzymatic activities. SIRT4 binds and represses GDH activity via ADP-ribosylation. SIRT5 deacetylates and activates carbamoyl phosphate synthetase 1 (CPS1), the rate-limiting step of the urea cycle.
Figure 6
SIRT1 plays key roles in cell survival and apoptosis. The complexity of SIRT1 regulation has made it a challenge to decipher SIRT1’s role in cancer. The SIRT1 gene is under the control of both environmental stimuli such as fasting and exercise and microRNAs (miRNAs), tenovins, and the hypermethylated in cancer 1 (HIC1) transcriptional repressor. The SIRT1 enzyme can also be modulated by protein-protein interactions with deleted in breast cancer 1 (DBC1), adaptor response to oxidative stress (AROS), Dif1, and necdin. SIRT1 interacts with and deacetylates numerous proteins involved in cell survival, DNA repair, and apoptosis (Figure 3). On the basis of this information, it has been difficult to predict what SIRT1 overexpression or activation by SIRT1-activating compounds will do in vivo, but so far experiments (171) point to SIRT1 acting as a tumor suppressor in the case of the p53−/+, breast cancer 1 (BRCA1), 7,12-dimethylbenz[α]anthracene (DMBA), and _APC_min+/− models of lymphoma, breast, skin, and colon cancers, respectively. Abbreviations: CR, calorie restriction; DBC1, deleted in breast cancer 1; FOXO, forkhead box, subgroup O; HIC1, hypermethylated in cancer 1; NF-κB, nuclear factor kappa B.
Figure 7
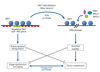
The relocalization of chromatin modifiers (RCM) hypothesis of aging stems from S. Imai and H. Kitano, who proposed in 1998 that changes in heterochromatin underlie the aging process. The idea was based in part on observations that, in response to DNA damage and aging, yeast SIR2 is released from silent loci and relocalized to DNA breaks, where it is hypothesized to organize chromatin to facilitate repair. During relocalization, expression of silent mating-type genes (HML and HMR) cause sterility, a hallmark of aging. Recent work shows that a similar process may drive aging in mammals. In response to DNA breaks or aging, SIRT1 also relocalizes away from open reading frames (ORFs) to DNA-break sites, seemingly to alter chromatin around the break site and recruit DNA damage–repair proteins such as RAD51 and NBS1. This relocalization of SIRT1, or the epigenetic changes it induces, is proposed to alter gene-expression patterns that result in tissue dysfunction and diseases associated with aging.
Figure 8
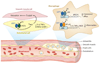
The role of SIRT1 in protection from atherosclerosis and cardiovascular disease (CVD). In the normal progression of atherosclerosis, damage and inflammation of blood vessel walls promote the infiltration of macrophages, where they take up and accumulate oxidized low-density lipoprotein, becoming foam cells that can eventually rupture and promote additional inflammation and plaque formation, which in turn occludes blood flow. Overexpression of SIRT1 in endothelial cells or treatment of mice with resveratrol SIRT1 reduces reactive oxygen species in vessel walls and slows the progression of CVD, apparently through multiple mechanisms. These include the suppression of inflammation by increasing endothelial nitric oxide synthase (eNOS) and decreasing nuclear factor kappa B (NF-κB) activity. Increased retrograde cholesterol transport in macrophages may also be a contributing factor. Abbreviations: cGMP, cyclic guanosine monophosphate; ICAM, intercellular adhesion molecule 1; IL-6, interleukin-6; iNOS, inducible NO, nitric oxide synthase; TNFα, tumor necrosis factor alpha.
Similar articles
- The sirtuin family's role in aging and age-associated pathologies.
Hall JA, Dominy JE, Lee Y, Puigserver P. Hall JA, et al. J Clin Invest. 2013 Mar;123(3):973-9. doi: 10.1172/JCI64094. Epub 2013 Mar 1. J Clin Invest. 2013. PMID: 23454760 Free PMC article. Review. - Sirtuins in aging and age-related disease.
Longo VD, Kennedy BK. Longo VD, et al. Cell. 2006 Jul 28;126(2):257-68. doi: 10.1016/j.cell.2006.07.002. Cell. 2006. PMID: 16873059 Review. - Recent progress in the biology and physiology of sirtuins.
Finkel T, Deng CX, Mostoslavsky R. Finkel T, et al. Nature. 2009 Jul 30;460(7255):587-91. doi: 10.1038/nature08197. Nature. 2009. PMID: 19641587 Free PMC article. Review. - SIRT1: linking adaptive cellular responses to aging-associated changes in organismal physiology.
Anastasiou D, Krek W. Anastasiou D, et al. Physiology (Bethesda). 2006 Dec;21:404-10. doi: 10.1152/physiol.00031.2006. Physiology (Bethesda). 2006. PMID: 17119153 Review. - Sirtuins and beta-cell function.
Bordone L, Guarente L. Bordone L, et al. Diabetes Obes Metab. 2007 Nov;9 Suppl 2:23-7. doi: 10.1111/j.1463-1326.2007.00769.x. Diabetes Obes Metab. 2007. PMID: 17919175
Cited by
- Unraveling Mitochondrial Reactive Oxygen Species Involvement in Psoriasis: The Promise of Antioxidant Therapies.
Ahmad Jamil H, Abdul Karim N. Ahmad Jamil H, et al. Antioxidants (Basel). 2024 Oct 11;13(10):1222. doi: 10.3390/antiox13101222. Antioxidants (Basel). 2024. PMID: 39456475 Free PMC article. Review. - Insights into Lysine Deacetylation of Natively Folded Substrate Proteins by Sirtuins.
Knyphausen P, de Boor S, Kuhlmann N, Scislowski L, Extra A, Baldus L, Schacherl M, Baumann U, Neundorf I, Lammers M. Knyphausen P, et al. J Biol Chem. 2016 Jul 8;291(28):14677-94. doi: 10.1074/jbc.M116.726307. Epub 2016 May 18. J Biol Chem. 2016. PMID: 27226597 Free PMC article. - SIRT7 inactivation reverses metastatic phenotypes in epithelial and mesenchymal tumors.
Malik S, Villanova L, Tanaka S, Aonuma M, Roy N, Berber E, Pollack JR, Michishita-Kioi E, Chua KF. Malik S, et al. Sci Rep. 2015 Apr 29;5:9841. doi: 10.1038/srep09841. Sci Rep. 2015. PMID: 25923013 Free PMC article. - SIRT1 and SIRT2: emerging targets in neurodegeneration.
Donmez G, Outeiro TF. Donmez G, et al. EMBO Mol Med. 2013 Mar;5(3):344-52. doi: 10.1002/emmm.201302451. Epub 2013 Feb 18. EMBO Mol Med. 2013. PMID: 23417962 Free PMC article. Review. - Saccharomyces boulardii modulates necrotizing enterocolitis in neonatal mice by regulating the sirtuin 1/NF‑κB pathway and the intestinal microbiota.
Zhang K, Zhang X, Lv A, Fan S, Zhang J. Zhang K, et al. Mol Med Rep. 2020 Aug;22(2):671-680. doi: 10.3892/mmr.2020.11138. Epub 2020 May 7. Mol Med Rep. 2020. PMID: 32626966 Free PMC article.
References
- Sinclair DA, Guarente L. Extrachromosomal rDNA circles—a cause of aging in yeast. Cell. 1997;91:1033–1042. - PubMed
- Sinclair DA, Guarente L. Unlocking the secrets of longevity genes. Sci. Am. 2006;294:48–57. - PubMed
- Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. - PubMed
- Westphal CH, Dipp MA, Guarente L. A therapeutic role for sirtuins in diseases of aging? Trends Biochem. Sci. 2007;32:555–560. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 AG027916-030003/AG/NIA NIH HHS/United States
- P01 AG027916-020003/AG/NIA NIH HHS/United States
- P01 AG027916-010003/AG/NIA NIH HHS/United States
- P01 AG027916-04S20003/AG/NIA NIH HHS/United States
- R01 AG028730/AG/NIA NIH HHS/United States
- R01 AG019719-07/AG/NIA NIH HHS/United States
- R01 AG028730-02/AG/NIA NIH HHS/United States
- R01 AG028730-01A1/AG/NIA NIH HHS/United States
- R01 AG019719/AG/NIA NIH HHS/United States
- R01 AG019719-06A1/AG/NIA NIH HHS/United States
- R01 AG028730-03/AG/NIA NIH HHS/United States
- P01 AG027916/AG/NIA NIH HHS/United States
- P01 AG027916-04S10003/AG/NIA NIH HHS/United States
- R01 AG032375/AG/NIA NIH HHS/United States
- P01 AG027916-040003/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous