Long-range gene regulation links genomic type 2 diabetes and obesity risk regions to HHEX, SOX4, and IRX3 - PubMed (original) (raw)
. 2010 Jan 12;107(2):775-80.
doi: 10.1073/pnas.0911591107. Epub 2009 Dec 22.
Enrico Moro, David Fredman, Pavla Navratilova, Øyvind Drivenes, Pär G Engström, M Eva Alonso, Elisa de la Calle Mustienes, José Luis Gómez Skarmeta, Maria J Tavares, Fernando Casares, Miguel Manzanares, Veronica van Heyningen, Anders Molven, Pål R Njølstad, Francesco Argenton, Boris Lenhard, Thomas S Becker
Affiliations
- PMID: 20080751
- PMCID: PMC2818943
- DOI: 10.1073/pnas.0911591107
Long-range gene regulation links genomic type 2 diabetes and obesity risk regions to HHEX, SOX4, and IRX3
Anja Ragvin et al. Proc Natl Acad Sci U S A. 2010.
Erratum in
- Proc Natl Acad Sci U S A. 2011 Mar 8;108(10):4264
Abstract
Genome-wide association studies identified noncoding SNPs associated with type 2 diabetes and obesity in linkage disequilibrium (LD) blocks encompassing HHEX-IDE and introns of CDKAL1 and FTO [Sladek R, et al. (2007) Nature 445:881-885; Steinthorsdottir V, et al. (2007) Nat. Genet 39:770-775; Frayling TM, et al. (2007) Science 316:889-894]. We show that these LD blocks contain highly conserved noncoding elements and overlap with the genomic regulatory blocks of the transcription factor genes HHEX, SOX4, and IRX3. We report that human highly conserved noncoding elements in LD with the risk SNPs drive expression in endoderm or pancreas in transgenic mice and zebrafish. Both HHEX and SOX4 have recently been implicated in pancreas development and the regulation of insulin secretion, but IRX3 had no prior association with pancreatic function or development. Knockdown of its orthologue in zebrafish, irx3a, increased the number of pancreatic ghrelin-producing epsilon cells and decreased the number of insulin-producing beta-cells and glucagon-producing alpha-cells, thereby suggesting a direct link of pancreatic IRX3 function to both obesity and type 2 diabetes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
Disease-associated SNPs (Top) and tested elements (elements 1–6) in the context of vertebrate GRBs around HHEX (A), SOX4 (B), and IRX3 (C). GRBs were defined by minimal synteny blocks to zebrafish (tan shading) or stickleback (light blue shading). Hapmap recombination hotspots (blue bars) mark the edges of LD blocks (blocks of interest shaded red). HCNE density plots for pairwise comparisons are colored yellow, orange, and red, based on the minimum percentage identity of the elements ≥50 bp in size used to derive them: human vs. mouse (mm: ≥95%, ≥98%, 100% identity), chicken (gg: ≥90, ≥95, ≥98), Xenopus (xt: ≥70, ≥80, ≥90) and zebrafish (dr: ≥70, ≥80, ≥90). HCNE densities were calculated as number of bases in HCNEs in sliding windows of 300 kb, and curves for different species are drawn to the same scale. HCNEs (black bars) are shown at the lowest threshold for each pairwise comparison. All features, including the suggested regulatory target gene (red) and bystander genes (gray) are shown in human coordinates. Gene annotations were adapted from the UCSC Known Genes set.
Fig. 2.
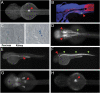
Reporter expression patterns directed by noncoding elements from T2D risk regions. (A and B) Live dorsal image (A) and cryosection (B) of a 72-hpf transgenic zebrafish showing GFP-reporter expression driven by the Xenopus element equivalent to the rs1111875-containing human HCNE downstream of HHEX (element 1). Red arrowheads point to the pancreatic anlagen. The horizontal line in (A) shows the level of section in B. The cryosection (B) was stained for DNA using DAPI (blue), for muscle actin using rhodamine-phalloidin (red) and for GFP using anti-GFP (green). (C) Sections through the pancreas of a 14.5-d mouse transgenic for rs1111875-containing human HCNE downstream of HHEX (element 1). LacZ staining is seen in the pancreatic anlagen. (D) Live dorsal image of a 48-hpf transgenic zebrafish showing GFP-reporter expression driven by the human HCNE closest to rs7754840 in CDKAL1 (element 2). Arrowheads point to expression in the hindbrain (red) and the primordium of the swim bladder (green). (E and F). Live lateral images of 48-hpf transgenic zebrafish showing GFP-reporter expression driven by the most deeply conserved HCNEs in the obesity-associated LD block of FTO. Element 3 (E) drives expression in the pronephric duct (red arrowhead), whereas element 4 (F) drives expression in the notochord (green arrowheads) and in hindbrain rhombomeres (red arrowhead). (G and H) Live dorsal images of 48-hpf transgenic zebrafish showing GFP-reporter expression driven by the rs1421085-containing HCNE and the rs9939609- containing HCNE in the FTO intron 1. Red arrowheads point to expression in the pancreatic area.
Fig. 3.
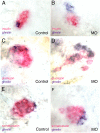
Pancreatic buds of 48-hpf zebrafish embryos showing a reduction in the number of insulin-expressing cells and an increase of ghrelin-expressing cells in irx3a morphants. In situ hybridization with insulin (red) and ghrelin (blue) probes on control (A) and irx3a morphant embryos (B). Statistical support for the observation is given in Table 3. (C and D) irx3a morphants display a reduction in pancreatic α-cells. In situ hybridization with glucagon (red in C and D), somatostatin (red in E and F), and ghrelin (blue in C_–_F) probes on control (C and E) and irx3a morphant embryos (D and F). showing reduction in the number of α-cells (D) and increase of ghrelin-expressing cells (D and F) in the pancreatic bud of 48-hpf irx3a morphant embryos. The number of δ-cells is unaffected. Embryos are in ventral view. Statistical support for the observation is given in Table 4.
Similar articles
- Obesity-associated variants within FTO form long-range functional connections with IRX3.
Smemo S, Tena JJ, Kim KH, Gamazon ER, Sakabe NJ, Gómez-Marín C, Aneas I, Credidio FL, Sobreira DR, Wasserman NF, Lee JH, Puviindran V, Tam D, Shen M, Son JE, Vakili NA, Sung HK, Naranjo S, Acemel RD, Manzanares M, Nagy A, Cox NJ, Hui CC, Gomez-Skarmeta JL, Nóbrega MA. Smemo S, et al. Nature. 2014 Mar 20;507(7492):371-5. doi: 10.1038/nature13138. Epub 2014 Mar 12. Nature. 2014. PMID: 24646999 Free PMC article. - BAC transgenic zebrafish reveal hypothalamic enhancer activity around obesity associated SNP rs9939609 within the human FTO gene.
Rinkwitz S, Geng FS, Manning E, Suster M, Kawakami K, Becker TS. Rinkwitz S, et al. Genesis. 2015 Oct;53(10):640-51. doi: 10.1002/dvg.22884. Epub 2015 Sep 5. Genesis. 2015. PMID: 26271004 Free PMC article. - Variations in the HHEX gene are associated with increased risk of type 2 diabetes in the Japanese population.
Horikoshi M, Hara K, Ito C, Shojima N, Nagai R, Ueki K, Froguel P, Kadowaki T. Horikoshi M, et al. Diabetologia. 2007 Dec;50(12):2461-6. doi: 10.1007/s00125-007-0827-5. Epub 2007 Oct 10. Diabetologia. 2007. PMID: 17928989 - Genetic origins of low birth weight.
Yaghootkar H, Freathy RM. Yaghootkar H, et al. Curr Opin Clin Nutr Metab Care. 2012 May;15(3):258-64. doi: 10.1097/MCO.0b013e328351f543. Curr Opin Clin Nutr Metab Care. 2012. PMID: 22406741 Review. - The role of the FTO (Fat Mass and Obesity Related) locus in regulating body size and composition.
Yeo GS. Yeo GS. Mol Cell Endocrinol. 2014 Nov;397(1-2):34-41. doi: 10.1016/j.mce.2014.09.012. Epub 2014 Sep 16. Mol Cell Endocrinol. 2014. PMID: 25224490 Review.
Cited by
- The fat mass and obesity-associated (FTO) gene allele rs9939609 and glucose tolerance, hepatic and total insulin sensitivity, in adults with obesity.
de Soysa AKH, Langaas M, Jakic A, Shojaee-Moradie F, Umpleby AM, Grill V, Mostad IL. de Soysa AKH, et al. PLoS One. 2021 Mar 8;16(3):e0248247. doi: 10.1371/journal.pone.0248247. eCollection 2021. PLoS One. 2021. PMID: 33684170 Free PMC article. Clinical Trial. - Genetics: Closing the distance on obesity culprits.
Gorkin DU, Ren B. Gorkin DU, et al. Nature. 2014 Mar 20;507(7492):309-10. doi: 10.1038/nature13212. Epub 2014 Mar 12. Nature. 2014. PMID: 24646989 No abstract available. - Interpreting human genetic variation with in vivo zebrafish assays.
Davis EE, Frangakis S, Katsanis N. Davis EE, et al. Biochim Biophys Acta. 2014 Oct;1842(10):1960-1970. doi: 10.1016/j.bbadis.2014.05.024. Epub 2014 Jun 2. Biochim Biophys Acta. 2014. PMID: 24887202 Free PMC article. Review. - Skeletal Muscle Immunometabolism in Women With Polycystic Ovary Syndrome: A Meta-Analysis.
Manti M, Stener-Victorin E, Benrick A. Manti M, et al. Front Physiol. 2020 Oct 22;11:573505. doi: 10.3389/fphys.2020.573505. eCollection 2020. Front Physiol. 2020. PMID: 33192572 Free PMC article. - Interaction analysis of FTO and IRX3 genes with obesity and related metabolic disorders in an admixed Latin American population: a possible risk increases of body weight excess.
Ruiz-Díaz MS, Mena-Yi D, Gómez-Camargo D, Mora-García G. Ruiz-Díaz MS, et al. Colomb Med (Cali). 2022 Jun 14;53(2):e2044874. doi: 10.25100/cm.v53i2.4874. eCollection 2022 Apr-Jun. Colomb Med (Cali). 2022. PMID: 36415696 Free PMC article.
References
- Sladek R, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–885. - PubMed
- Saxena R, et al. Diabetes Genetics Initiative of Broad Institute of Harvard and MIT, Lund University, and Novartis Institutes of BioMedical Research. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. - PubMed
- Steinthorsdottir V, et al. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat Genet. 2007;39:770–775. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials