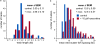Replication factory activation can be decoupled from the replication timing program by modulating Cdk levels - PubMed (original) (raw)
Replication factory activation can be decoupled from the replication timing program by modulating Cdk levels
Alexander M Thomson et al. J Cell Biol. 2010.
Abstract
In the metazoan replication timing program, clusters of replication origins located in different subchromosomal domains fire at different times during S phase. We have used Xenopus laevis egg extracts to drive an accelerated replication timing program in mammalian nuclei. Although replicative stress caused checkpoint-induced slowing of the timing program, inhibition of checkpoint kinases in an unperturbed S phase did not accelerate it. Lowering cyclin-dependent kinase (Cdk) activity slowed both replication rate and progression through the timing program, whereas raising Cdk activity increased them. Surprisingly, modest alteration of Cdk activity changed the amount of DNA synthesized during different stages of the timing program. This was associated with a change in the number of active replication factories, whereas the distribution of origins within active factories remained relatively normal. The ability of Cdks to differentially effect replication initiation, factory activation, and progression through the timing program provides new insights into the way that chromosomal DNA replication is organized during S phase.
Figures
Figure 1.
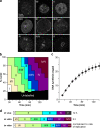
Progression of CHOC-400 nuclei through the timing program in vitro. CHOC-400 nuclei were incubated at 10,000 nuclei/µl (∼60 ng DNA/µl) in egg extracts supplemented with geminin. (a) Representative images of CHO nuclei incubated in X. laevis egg extracts and pulse labeled for 5 min with Cy3-dUTP. See Materials and methods for full description of the different labeling patterns. (b) At different times, aliquots were pulse labeled for 5 min with Cy3-dUTP, and the proportion of different replication patterns was assessed. (c) Extract was supplemented with α-[32P]dATP. At different times, total DNA synthesis was measured by TCA precipitation and scintillation counting. The SEM of 14 independent experiments is shown. (d, top) The time of appearance of replication patterns during S phase of CHOC-400 cells as described previously by Dimitrova and Gilbert (1999). (middle) The proportion of time spent by CHOC-400 nuclei replicating specific patterns as calculated in b. (bottom) The amount of DNA synthesis associated with each replication pattern in vitro calculated by scaling the proportions in b by the rate of replication (c).
Figure 2.
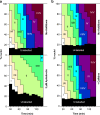
Initiation times associated with different replication patterns. CHOC-400 nuclei were incubated at 10,000 nuclei/µl in egg extracts supplemented with geminin. At different times, aliquots were supplemented with 1 mM roscovitine to block further initiation events. (a) Extract was also supplemented with α-[32P]dATP at the start of the incubation. At different times thereafter, total DNA synthesis was measured by TCA precipitation and scintillation counting. (b) At different times, extract was pulse labeled with Cy3-dUTP for 5 min, and the proportion of nuclei showing different replication patterns was assessed. (c) Data from a and b were combined to provide an estimate of the times over which most initiation events associated with the different replication patterns were occurring.
Figure 3.
Effect of aphidicolin and caffeine on the timing program. (a and b) CHOC-400 nuclei were incubated at 10,000 nuclei/µl in egg extracts supplemented with geminin ± 3 µM aphidicolin (a) or 5 mM caffeine (b). At different times, extract was pulse labeled with Cy3-dUTP for 5 min, and the proportion of nuclei showing different replication patterns was assessed.
Figure 4.
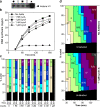
Effect of low concentrations of roscovitine on the timing program. (a) Extract was supplemented with histone H1, γ-[32P]ATP, and various concentrations of roscovitine. After incubation for 90 min, protein was separated by SDS-PAGE, and 32P incorporation into H1 was assessed by phosphorimager. (b and c) CHOC-400 nuclei were incubated at 10,000 nuclei/µl in egg extracts supplemented with geminin and different concentrations of roscovitine. Aliquots were also supplemented with α-[32P]dATP at the start of the incubation. (b) At different times thereafter, total DNA synthesis was measured by TCA precipitation and scintillation counting. (c) At different times, extract was pulse labeled with Cy3-dUTP for 5 min, and the proportion of nuclei showing different replication patterns was assessed. No replication patterns were visible in extracts treated with 300 µM roscovitine.
Figure 5.
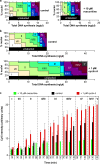
Effect of recombinant cyclin A on the timing program. (a) Extract was supplemented with histone H1 γ-[32P]ATP and various concentrations of cyclin A. After incubation for 90 min, protein was separated by SDS-PAGE and 32P incorporation into H1 assessed by phosphorimager. (b and c) CHOC-400 nuclei were incubated at 10,000 nuclei/µl in egg extracts supplemented with geminin and different concentrations of recombinant cyclin A. (b) Aliquots were also supplemented with α-[32P]dATP at the start of the incubation. At different times afterward, total DNA synthesis was measured by TCA precipitation and scintillation counting. (c) At either 45 or 85 min, extract was pulse labeled with Cy3-dUTP for 5 min, and the proportion of nuclei showing different replication patterns was assessed. (d) Extract was supplemented ± 1 pM cyclin A. At different times, extract was pulse labeled with Cy3-dUTP for 5 min, and the proportion of nuclei showing different replication patterns was assessed.
Figure 6.
Decoupling of replication and the timing program. CHOC-400 nuclei were incubated at 10,000 nuclei/µl in egg extracts supplemented with geminin ± 10 µM roscovitine or 1 pM recombinant cyclin A. (a and b) Data from Fig. 5 and
Fig. S2
are shown where the replication patterns at different times are plotted against the total DNA synthesis that had occurred at each time. Vertical ticks above the plot indicate the boundaries of different time points. (c) Extract was also supplemented with Cy5-dCTP at the start of the incubation. At different times, aliquots were pulse labeled with Cy3-dUTP for 5 min, and individual nuclei were analyzed for replication pattern (Cy3 label) or total DNA synthesis (Cy5 label). Error bars indicate mean ± SD.
Figure 7.
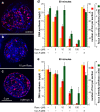
Replication foci number depends on Cdk levels. CHOC-400 nuclei were incubated in X. laevis egg extracts incubated at 10,000 nuclei/µl in egg extracts supplemented with the indicated concentrations of roscovitine (rosc) or cyclin A (cyc A). Parallel incubations were supplemented with α-[32P]dATP to measure total DNA synthesis. At 50 or 90 min, extract was pulsed for 5 min with Cy3-dUTP. (a–c) Representative images from the 50-min time point are shown. (d and e) The number and mean intensity of Cy3-labeled foci at 50 (d) and 90 (e) min were measured. Error bars indicate SEM between different nuclei.
Figure 8.
DNA fiber analysis. (a and b) CHOC-400 nuclei were incubated at 10,000 nuclei/µl in egg extracts supplemented with geminin minus (blue) or plus (red) 10 µM roscovitine (rosc). After incubation for 45 min, extract was pulsed with BrdUTP for 5 min. DNA was isolated, spread on glass slides, and the BrdU-labeled tracks were analyzed. The length of isolated tracks (a) and the distribution of forks within clusters are shown (b).
Figure 9.
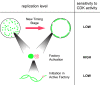
Cdk sensitivity of different levels of replication control. Cartoon showing three different levels of S phase control. The top level shows progression between two different stages of the timing program for a single nucleus. Small green dots represent replication factories, and the black circles represent the nuclear envelope. The middle level shows the activation of a new replication factory (pink dots) next to an existing factory (large green dot). The lower level shows the initiation of a new replication origin (pink circle) on a strand of DNA (green) within an active factory (active replicon cluster).
Similar articles
- Clusters, factories and domains: The complex structure of S-phase comes into focus.
Gillespie PJ, Blow JJ. Gillespie PJ, et al. Cell Cycle. 2010 Aug 15;9(16):3218-26. doi: 10.4161/cc.9.16.12644. Epub 2010 Aug 11. Cell Cycle. 2010. PMID: 20724827 Free PMC article. - DNA replication of mitotic chromatin in Xenopus egg extracts.
Prokhorova TA, Mowrer K, Gilbert CH, Walter JC. Prokhorova TA, et al. Proc Natl Acad Sci U S A. 2003 Nov 11;100(23):13241-6. doi: 10.1073/pnas.2336104100. Epub 2003 Nov 3. Proc Natl Acad Sci U S A. 2003. PMID: 14597706 Free PMC article. - Association of cyclin A and cdk2 with SV40 DNA in replication initiation complexes is cell cycle dependent.
Cannella D, Roberts JM, Fotedar R. Cannella D, et al. Chromosoma. 1997 Apr;105(6):349-59. doi: 10.1007/BF02529750. Chromosoma. 1997. PMID: 9087377 - WEE1 kinase limits CDK activities to safeguard DNA replication and mitotic entry.
Elbæk CR, Petrosius V, Sørensen CS. Elbæk CR, et al. Mutat Res. 2020 Jan-Apr;819-820:111694. doi: 10.1016/j.mrfmmm.2020.111694. Epub 2020 Feb 25. Mutat Res. 2020. PMID: 32120135 Review. - A quantitative model for the cdc2 control of S phase and mitosis in fission yeast.
Stern B, Nurse P. Stern B, et al. Trends Genet. 1996 Sep;12(9):345-50. Trends Genet. 1996. PMID: 8855663 Review.
Cited by
- The role of DDK and Treslin-MTBP in coordinating replication licensing and pre-initiation complex formation.
Volpi I, Gillespie PJ, Chadha GS, Blow JJ. Volpi I, et al. Open Biol. 2021 Oct;11(10):210121. doi: 10.1098/rsob.210121. Epub 2021 Oct 27. Open Biol. 2021. PMID: 34699733 Free PMC article. - The Ph1 locus suppresses Cdk2-type activity during premeiosis and meiosis in wheat.
Greer E, Martín AC, Pendle A, Colas I, Jones AM, Moore G, Shaw P. Greer E, et al. Plant Cell. 2012 Jan;24(1):152-62. doi: 10.1105/tpc.111.094771. Epub 2012 Jan 24. Plant Cell. 2012. PMID: 22274628 Free PMC article. - Uncovering the role of APC-Cdh1 in generating the dynamics of S-phase onset.
Yuan X, Srividhya J, De Luca T, Lee JH, Pomerening JR. Yuan X, et al. Mol Biol Cell. 2014 Feb;25(4):441-56. doi: 10.1091/mbc.E13-08-0480. Epub 2013 Dec 19. Mol Biol Cell. 2014. PMID: 24356446 Free PMC article. - Organization of DNA Replication Origin Firing in Xenopus Egg Extracts: The Role of Intra-S Checkpoint.
Ciardo D, Haccard O, Narassimprakash H, Arbona JM, Hyrien O, Audit B, Marheineke K, Goldar A. Ciardo D, et al. Genes (Basel). 2021 Aug 9;12(8):1224. doi: 10.3390/genes12081224. Genes (Basel). 2021. PMID: 34440398 Free PMC article. - Overexpression of hepatocyte nuclear factor-4α initiates cell cycle entry, but is not sufficient to promote β-cell expansion in human islets.
Rieck S, Zhang J, Li Z, Liu C, Naji A, Takane KK, Fiaschi-Taesch NM, Stewart AF, Kushner JA, Kaestner KH. Rieck S, et al. Mol Endocrinol. 2012 Sep;26(9):1590-602. doi: 10.1210/me.2012-1019. Epub 2012 Jul 13. Mol Endocrinol. 2012. PMID: 22798294 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- C303/A7399/CRUK_/Cancer Research UK/United Kingdom
- A5434/CRUK_/Cancer Research UK/United Kingdom
- BBS/S/P/2003/10310/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- A3135/CRUK_/Cancer Research UK/United Kingdom
- C303/A3135/CRUK_/Cancer Research UK/United Kingdom
- A7399/CRUK_/Cancer Research UK/United Kingdom
- C303/A5434/CRUK_/Cancer Research UK/United Kingdom
LinkOut - more resources
Full Text Sources