Selective inhibitors of the JMJD2 histone demethylases: combined nondenaturing mass spectrometric screening and crystallographic approaches - PubMed (original) (raw)
. 2010 Feb 25;53(4):1810-8.
doi: 10.1021/jm901680b.
Affiliations
- PMID: 20088513
- PMCID: PMC2825117
- DOI: 10.1021/jm901680b
Free PMC article
Selective inhibitors of the JMJD2 histone demethylases: combined nondenaturing mass spectrometric screening and crystallographic approaches
Nathan R Rose et al. J Med Chem. 2010.
Free PMC article
Abstract
Ferrous ion and 2-oxoglutarate (2OG) oxygenases catalyze the demethylation of N(epsilon)-methylated lysine residues in histones. Here we report studies on the inhibition of the JMJD2 subfamily of histone demethylases, employing binding analyses by nondenaturing mass spectrometry (MS), dynamic combinatorial chemistry coupled to MS, turnover assays, and crystallography. The results of initial binding and inhibition assays directed the production and analysis of a set of N-oxalyl-d-tyrosine derivatives to explore the extent of a subpocket at the JMJD2 active site. Some of the inhibitors were shown to be selective for JMJD2 over the hypoxia-inducible factor prolyl hydroxylase PHD2. A crystal structure of JMJD2A in complex with one of the potent inhibitors was obtained; modeling other inhibitors based on this structure predicts interactions that enable improved inhibition for some compounds.
Figures
Figure 1
Reactions catalyzed by the JMJD2 histone demethylases. Each step is coupled to the conversion of O2 and 2-oxoglutarate to succinate and CO2. 2OG = 2-oxoglutarate, succ = succinate.
Figure 2
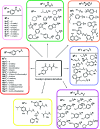
Structures of potential inhibitors used in this study.
Figure 3
Dynamic combinatorial mass spectrometry (DCMS) approach. (A) The _N_-oxalyl group of support ligand 1b anchors the molecule into the active site of JMJD2E via interaction with the Fe(II) ion (magenta), leaving the thiol side chain free for disulfide formation with the thiol library in solution. (B) Selective formation of a JMJD2E−disulfide complex with the thiol member that fits best into the active site.
Figure 4
Structures of the _N_-oxalylamino acids investigated in this study.
Figure 5
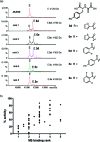
Correlation of nondenaturing ESI-MS analyses and inhibition results for JMJD2E inhibitors. (a) Compounds were grouped in five ranking sets reflecting the strength of binding to the JMJD2E·Fe(II)·Zn(II) complex (E). Rank 1: ∼1:4 ratio unbound:bound; rank 2: ∼1:2 unbound:bound; rank 3: ∼1:1 unbound:bound; rank 4: ∼4:1 unbound:bound, and rank 5: ∼10:1 unbound:bound. The MS spectra show examples of data for representative compounds from each ranking set. Some samples (11 of the 73 compounds tested) were not considered to produce spectra of sufficient quality for classification and were thus excluded from the ranking. (b) Initial rates of all compounds tested as JMJD2E inhibitors (100 μM) binding rank as determined by ESI-MS, demonstrating correlation between the two data sets. Kendall’s τB = 0.58 (p < 0.0001), Spearman’s ρ = 0.72 (p < 0.0001).
Figure 6
Screening for JMJD2E inhibitors. Compounds were tested as JMJD2E inhibitors (100 μM) using the FDH assay, and the percentage activities relative to the DMSO control are shown. Measurements were made in duplicate and shown as averages with standard errors of the mean.
Figure 7
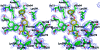
Stereoviews from the JMJD2A:10a crystal structure. (a) Stereoview of the crystal structure of JMJD2A (transparent ribbons and green sticks) in complex with compound 10a (yellow sticks). Ni(II) (orange sphere) replaces Fe(II) in the active site; the structural Zn(II) is shown as a blue sphere. Highlighted residues (green) chelate active site metal and form part of the substrate/cosubstrate binding pocket. (b) Close-up stereoview of substrate/cosubstrate binding site with compound 10a, showing details of hydrogen bonding interactions between 10a, Lys206 and Tyr132 side chains, and the backbone amide nitrogen of Ala186.
Figure 8
Electron density map (stereoview) showing compound 10a (yellow sticks) in the active site of JMJD2A (green sticks). The experimental 2_F_o − _F_c electron density, displayed as blue mesh, is shown for the metal (gray sphere) coordinating residues and ligands (contoured to 1σ).
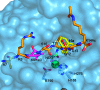
Figure 9
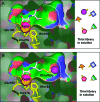
Binding of 10a to JMJD2A likely interferes with both 2OG and histone substrate binding. Superimposition of H3K9me3 substrate (orange and pink sticks) with JMJD2A·Ni(II)·Zn(II).10a structure (PDB ID 2OQ6) is shown. The surface of JMJD2A (blue) illustrates separate K9me3 (pink) and cosubstrate (NOG, cyan, replaces 2OG in this structure) subpockets. Compound 10a (yellow sticks) occupies a large hydrophobic pocket adjacent to the substrate binding cleft. It may interfere with binding of the Thr11 side chain of the H3K9 substrate.
Similar articles
- Inhibition of the histone lysine demethylase JMJD2A by ejection of structural Zn(II).
Sekirnik R, Rose NR, Thalhammer A, Seden PT, Mecinović J, Schofield CJ. Sekirnik R, et al. Chem Commun (Camb). 2009 Nov 14;(42):6376-8. doi: 10.1039/b916357c. Epub 2009 Sep 28. Chem Commun (Camb). 2009. PMID: 19841782 - Inhibition of the histone demethylase JMJD2E by 3-substituted pyridine 2,4-dicarboxylates.
Thalhammer A, Mecinović J, Loenarz C, Tumber A, Rose NR, Heightman TD, Schofield CJ. Thalhammer A, et al. Org Biomol Chem. 2011 Jan 7;9(1):127-35. doi: 10.1039/c0ob00592d. Epub 2010 Nov 15. Org Biomol Chem. 2011. PMID: 21076780 Free PMC article. - Structural investigations of the nickel-induced inhibition of truncated constructs of the JMJD2 family of histone demethylases using X-ray absorption spectroscopy.
Giri NC, Passantino L, Sun H, Zoroddu MA, Costa M, Maroney MJ. Giri NC, et al. Biochemistry. 2013 Jun 18;52(24):4168-83. doi: 10.1021/bi400274v. Epub 2013 Jun 7. Biochemistry. 2013. PMID: 23692052 Free PMC article. - Structure-function relationships in KDM7 histone demethylases.
Chaturvedi SS, Ramanan R, Waheed SO, Karabencheva-Christova TG, Christov CZ. Chaturvedi SS, et al. Adv Protein Chem Struct Biol. 2019;117:113-125. doi: 10.1016/bs.apcsb.2019.08.005. Epub 2019 Sep 10. Adv Protein Chem Struct Biol. 2019. PMID: 31564306 Review. - KDM4/JMJD2 histone demethylases: epigenetic regulators in cancer cells.
Berry WL, Janknecht R. Berry WL, et al. Cancer Res. 2013 May 15;73(10):2936-42. doi: 10.1158/0008-5472.CAN-12-4300. Epub 2013 May 3. Cancer Res. 2013. PMID: 23644528 Free PMC article. Review.
Cited by
- Hypoxia-inducible factor prolyl hydroxylase inhibition: robust new target or another big bust for stroke therapeutics?
Karuppagounder SS, Ratan RR. Karuppagounder SS, et al. J Cereb Blood Flow Metab. 2012 Jul;32(7):1347-61. doi: 10.1038/jcbfm.2012.28. Epub 2012 Mar 14. J Cereb Blood Flow Metab. 2012. PMID: 22415525 Free PMC article. Review. - Conservation of the unusual dimeric JmjC fold of JMJD7 from Drosophila melanogaster to humans.
Chowdhury R, Abboud MI, Wiley J, Tumber A, Markolovic S, Schofield CJ. Chowdhury R, et al. Sci Rep. 2022 Apr 11;12(1):6065. doi: 10.1038/s41598-022-10028-y. Sci Rep. 2022. PMID: 35410347 Free PMC article. - The Histone Demethylase KDM3A, Increased in Human Pancreatic Tumors, Regulates Expression of DCLK1 and Promotes Tumorigenesis in Mice.
Dandawate P, Ghosh C, Palaniyandi K, Paul S, Rawal S, Pradhan R, Sayed AAA, Choudhury S, Standing D, Subramaniam D, Padhye SB, Gunewardena S, Thomas SM, Neil MO, Tawfik O, Welch DR, Jensen RA, Maliski S, Weir S, Iwakuma T, Anant S, Dhar A. Dandawate P, et al. Gastroenterology. 2019 Dec;157(6):1646-1659.e11. doi: 10.1053/j.gastro.2019.08.018. Epub 2019 Aug 20. Gastroenterology. 2019. PMID: 31442435 Free PMC article. - KDM4B: A Nail for Every Hammer?
Wilson C, Krieg AJ. Wilson C, et al. Genes (Basel). 2019 Feb 12;10(2):134. doi: 10.3390/genes10020134. Genes (Basel). 2019. PMID: 30759871 Free PMC article. Review. - The role of histone demethylases in cancer therapy.
Hoffmann I, Roatsch M, Schmitt ML, Carlino L, Pippel M, Sippl W, Jung M. Hoffmann I, et al. Mol Oncol. 2012 Dec;6(6):683-703. doi: 10.1016/j.molonc.2012.07.004. Epub 2012 Aug 7. Mol Oncol. 2012. PMID: 22902149 Free PMC article. Review.
References
- Klose R. J.; Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat. Rev. Mol. Cell Biol. 2007, 8 (4), 307–318. - PubMed
- Tsukada Y.; Fang J.; Erdjument-Bromage H.; Warren M. E.; Borchers C. H.; Tempst P.; Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature 2006, 439 (7078), 811–816. - PubMed
- Cloos P. A.; Christensen J.; Agger K.; Maiolica A.; Rappsilber J.; Antal T.; Hansen K. H.; Helin K. The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature 2006, 442 (7100), 307–311. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information