Analysis of host-parasite incongruence in papillomavirus evolution using importance sampling - PubMed (original) (raw)
Analysis of host-parasite incongruence in papillomavirus evolution using importance sampling
Seena D Shah et al. Mol Biol Evol. 2010 Jun.
Abstract
The papillomaviruses (PVs) are a family of viruses infecting several mammalian and nonmammalian species that cause cervical cancer in humans. The evolutionary history of the PVs as it associated with a wide range of host species is not well understood. Incongruities between the phylogenetic trees of various viral genes as well as between these genes and the host phylogenies suggest historical viral recombination as well as violations of strict virus-host cospeciation. The extent of recombination events among PVs is uncertain, however, and there is little evidence to support a theory of PV spread via recent host transfers. We have investigated incongruence between PV genes and hence, the possibility of recombination, using Bayesian phylogenetic methods. We find significant evidence for phylogenetic incongruence among the six PV genes E1, E2, E6, E7, L1, and L2, indicating substantial recombination. Analysis of E1 and L1 phylogenies suggests ancestral recombination events. We also describe a new method for examining alternative host-parasite association mechanisms by applying importance sampling to Bayesian divergence time estimation. This new approach is not restricted by a fixed viral tree topology or knowledge of viral divergence times, multiple parasite taxa per host may be included, and it can distinguish between prior divergence of the virus before host speciation and host transfer of the virus following speciation. Using this method, we find prior divergence of PV lineages associated with the ancestral mammalian host resulting in at least 6 PV lineages prior to speciation of this host. These PV lineages have then followed paths of prior divergence and cospeciation to eventually become associated with the extant host species. Only one significant instance of host transfer is supported, the transfer of the ancestral L1 gene between a Primate and Hystricognathi host based on the divergence times between the upsilon human type 41 and porcupine PVs.
Figures
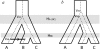
FIG. 1.
Viral diversification mechanisms that may result in incongruent topologies between virus (dashed) and host (solid). (a) A virus lineage associated with the ancestral host ABC cospeciates with its host at the initial speciation event. The descendent virus lineage in host BC is not observed in host B—this may be due to extinction of the virus in B, failure of the ancestral lineage virus to associate with B following speciation of host BC (“incomplete lineage sorting”), or a failure to detect the virus. The virus lineage that has been detected in B is the result of a host transfer of the virus lineage from A. This host transfer event can be identified by comparing the host and viral divergence times as VAB < HA(BC) in contrast to the previous node where VAC = HA(BC) consistent with cospeciation of the viruses in A and C with their hosts. (_b_) The virus lineage associated with ancestral host ABC diverges prior to the first host speciation event resulting in two virus lineages that can then segregate along the descendant hosts independently of each other. The viruses associated with hosts A and B are descended from one lineage, whereas the virus in host C is descended from the other lineage. Once again, these events are reflected in divergence times: VAB = HA(BC) as the virus cospeciated with its host, however, VAC > HA(BC) indicating the divergence of virus C prior to speciation of host ABC.
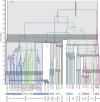
FIG. 2.
The E1 and L1 gene trees each shown on top of the associated host tree (grey), which is scaled according to the times of the host divergences (Ma). The timings of the PV splits correspond with the mean times sampled from the biased sampling analysis of each gene; the 95% CIs of viral divergence times at each node are represented by the colored bars. The host speciation times of related host taxa are highlighted in dark grey. Posterior probabilities of internal PV branches are indicated beside the branches. PV nodes labeled with a * indicate divergences for which cospeciation violations were found to be statistically significant. Labels below the tree indicate 1) the names of the PV taxa—“_α_-2 HPVs” groups together all HPVs included in our analysis from species 2 of the α genus, 2) the genus classifications of the PV taxa, 3) the host species from which the virus was isolated. PV clades are colored according to genus classifications; for simplicity, some genera that consistently group together in both gene trees have been assigned the same color.
FIG. 2.
The E1 and L1 gene trees each shown on top of the associated host tree (grey), which is scaled according to the times of the host divergences (Ma). The timings of the PV splits correspond with the mean times sampled from the biased sampling analysis of each gene; the 95% CIs of viral divergence times at each node are represented by the colored bars. The host speciation times of related host taxa are highlighted in dark grey. Posterior probabilities of internal PV branches are indicated beside the branches. PV nodes labeled with a * indicate divergences for which cospeciation violations were found to be statistically significant. Labels below the tree indicate 1) the names of the PV taxa—“_α_-2 HPVs” groups together all HPVs included in our analysis from species 2 of the α genus, 2) the genus classifications of the PV taxa, 3) the host species from which the virus was isolated. PV clades are colored according to genus classifications; for simplicity, some genera that consistently group together in both gene trees have been assigned the same color.
FIG. 3.
A splits network generated from the E1 and L1 consensus trees using SplitsTree. Sets of parallel edges in the network indicate locations of incongruence and potential recombination.
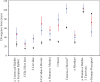
FIG. 4.
Divergence times for the host (black), E1 (red), and L1 (blue) genes. CIs for the host and viral divergence times are indicated with error bars; unseen error bars represent CIs smaller than the size of the symbols. Viral divergence times further back than host divergence times (e.g., human–monkey) represent prior divergence, whereas viral divergence times more recent than host divergence times (e.g., human–porcupine) represent likely host transfer events. †Statistically significant violation of host divergence time observed for E1 only. ‡Statistically significant violation of host divergence time observed for L1 only. *Node present in L1 gene tree only.
Similar articles
- Recombination Between High-Risk Human Papillomaviruses and Non-Human Primate Papillomaviruses: Evidence of Ancient Host Switching Among Alphapapillomaviruses.
Murahwa AT, Tshabalala M, Williamson AL. Murahwa AT, et al. J Mol Evol. 2020 Jul;88(5):453-462. doi: 10.1007/s00239-020-09946-0. Epub 2020 May 8. J Mol Evol. 2020. PMID: 32385625 Free PMC article. - Non-human Primate Papillomaviruses Share Similar Evolutionary Histories and Niche Adaptation as the Human Counterparts.
Chen Z, Long T, Wong PY, Ho WCS, Burk RD, Chan PKS. Chen Z, et al. Front Microbiol. 2019 Sep 10;10:2093. doi: 10.3389/fmicb.2019.02093. eCollection 2019. Front Microbiol. 2019. PMID: 31552003 Free PMC article. - Patterns of co-speciation and host switching in primate malaria parasites.
Garamszegi LZ. Garamszegi LZ. Malar J. 2009 May 22;8:110. doi: 10.1186/1475-2875-8-110. Malar J. 2009. PMID: 19463162 Free PMC article. - Adaptation of Alpha-Papillomavirus over Millennia.
de Oliveira CM. de Oliveira CM. Acta Cytol. 2019;63(2):97-99. doi: 10.1159/000492658. Epub 2018 Dec 13. Acta Cytol. 2019. PMID: 30544125 Review. - Papillomaviruses: different genes have different histories.
García-Vallvé S, Alonso A, Bravo IG. García-Vallvé S, et al. Trends Microbiol. 2005 Nov;13(11):514-21. doi: 10.1016/j.tim.2005.09.003. Epub 2005 Sep 21. Trends Microbiol. 2005. PMID: 16181783 Review.
Cited by
- Synonymous nucleotide changes drive papillomavirus evolution.
King KM, Rajadhyaksha EV, Tobey IG, Van Doorslaer K. King KM, et al. Tumour Virus Res. 2022 Dec;14:200248. doi: 10.1016/j.tvr.2022.200248. Epub 2022 Oct 17. Tumour Virus Res. 2022. PMID: 36265836 Free PMC article. Review. - Population genomics of the Wolbachia endosymbiont in Drosophila melanogaster.
Richardson MF, Weinert LA, Welch JJ, Linheiro RS, Magwire MM, Jiggins FM, Bergman CM. Richardson MF, et al. PLoS Genet. 2012;8(12):e1003129. doi: 10.1371/journal.pgen.1003129. Epub 2012 Dec 20. PLoS Genet. 2012. PMID: 23284297 Free PMC article. - New insights into Sauropsid Papillomaviridae evolution and epizootiology: discovery of two novel papillomaviruses in native and invasive Island geckos.
Agius JE, Phalen DN, Rose K, Eden JS. Agius JE, et al. Virus Evol. 2019 Nov 22;5(2):vez051. doi: 10.1093/ve/vez051. eCollection 2019 Jul. Virus Evol. 2019. PMID: 31798966 Free PMC article. Review. - Evolutionary dynamics of ten novel Gamma-PVs: insights from phylogenetic incongruence, recombination and phylodynamic analyses.
Murahwa AT, Nindo F, Onywera H, Meiring TL, Martin DP, Williamson AL. Murahwa AT, et al. BMC Genomics. 2019 May 14;20(1):368. doi: 10.1186/s12864-019-5735-9. BMC Genomics. 2019. PMID: 31088349 Free PMC article. - The Role of aDNA in Understanding the Coevolutionary Patterns of Human Sexually Transmitted Infections.
Pimenoff VN, Houldcroft CJ, Rifkin RF, Underdown S. Pimenoff VN, et al. Genes (Basel). 2018 Jun 25;9(7):317. doi: 10.3390/genes9070317. Genes (Basel). 2018. PMID: 29941858 Free PMC article. Review.
References
- Antonsson A, McMillan NA. Papillomavirus in healthy skin of Australian animals. J Gen Virol. 2006;87:3195–3200. - PubMed
- Bernard H-U. Coevolution of papillomaviruses with human populations. Trends Microbiol. 1994;2:140–143. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials