Modulation of heat shock transcription factor 1 as a therapeutic target for small molecule intervention in neurodegenerative disease - PubMed (original) (raw)
Modulation of heat shock transcription factor 1 as a therapeutic target for small molecule intervention in neurodegenerative disease
Daniel W Neef et al. PLoS Biol. 2010.
Abstract
Neurodegenerative diseases such as Huntington disease are devastating disorders with no therapeutic approaches to ameliorate the underlying protein misfolding defect inherent to poly-glutamine (polyQ) proteins. Given the mounting evidence that elevated levels of protein chaperones suppress polyQ protein misfolding, the master regulator of protein chaperone gene transcription, HSF1, is an attractive target for small molecule intervention. We describe a humanized yeast-based high-throughput screen to identify small molecule activators of human HSF1. This screen is insensitive to previously characterized activators of the heat shock response that have undesirable proteotoxic activity or that inhibit Hsp90, the central chaperone for cellular signaling and proliferation. A molecule identified in this screen, HSF1A, is structurally distinct from other characterized small molecule human HSF1 activators, activates HSF1 in mammalian and fly cells, elevates protein chaperone expression, ameliorates protein misfolding and cell death in polyQ-expressing neuronal precursor cells and protects against cytotoxicity in a fly model of polyQ-mediated neurodegeneration. In addition, we show that HSF1A interacts with components of the TRiC/CCT complex, suggesting a potentially novel regulatory role for this complex in modulating HSF1 activity. These studies describe a novel approach for the identification of new classes of pharmacological interventions for protein misfolding that underlies devastating neurodegenerative disease.
Conflict of interest statement
Duke University has filed a patent application for the screening technology described in this report as well as the small molecules identified via this screening technology.
Figures
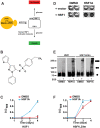
Figure 1. HSF1A activates human HSF1 function in yeast.
(A) Strategy used to identify chemical activators of human HSF1 in yeast. Yeast cells expressing the essential yeast HSF under control of the glucose-repressible GAL1 promoter are dependent on galactose for growth. Upon shifting cells to glucose-containing medium, the cells are dependent on activation of human HSF1 for growth. (B) Structure of HSF1A. (C) Yeast cells expressing wild-type human HSF1 were supplemented with 10 µM HSF1A or DMSO and grown in 96-well plates for 4 d. Growth was monitored by measuring OD600. (D) HSF1-dependence for HSF1A-mediated cell growth. Strain DNY75 expressing either human HSF1 (+HSF1) or an empty vector (+vector) were seeded into microtiter wells and incubated in the presence of HSF1A or DMSO solvent for 4 d and then photographed. Note that only cells expressing human HSF1 grow in response to HSF1A. (E) Yeast strain DNY75 was grown in the presence of DMSO or 20 µM HSF1A for 18 h, and HSF1 multimerization was evaluated by EGS cross-linking, SDS-PAGE, and immunoblotting. The positions of molecular weight markers are indicated on the left, and circles indicating the expected migration of HSF1 monomers, dimers, and trimers are on the right. (F) Yeast cells expressing HSF1LZ4m were supplemented with 10 µM HSF1A or DMSO and grown in 96-well plates for 4 d. Growth was monitored by measuring OD600.
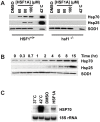
Figure 2. HSF1A activates Hsp70 expression in mammalian cells.
(A) HSF1+/+ and hsf1−/− MEFs were treated with DMSO solvent or increasing concentrations of HSF1A for 15 h or heat shocked for 2 h at 42°C followed by a 15-h recovery. Total protein was analyzed for Hsp70 and Hsp25 expression by immunoblotting. SOD1 serves as a loading control. (B) HSF1+/+ MEF cells were treated with 80 µM HSF1A over time and Hsp70 and Hsp25 levels analyzed by immunoblotting. (C) HSF1+/+ MEF cells were treated with 80 µM HSF1A for 6 h or heat shocked at 42°C for 2 h, and total RNA was analyzed by RNA blotting. 18S rRNA serves as a loading control.
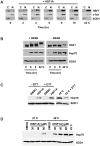
Figure 3. Features of HSF1A-dependent HSF1 activation.
(A) HSF1+/+ MEFs were treated with 80 µM HSF1A for the indicated time in hours or heat shocked at 42°C for 2 h, and nuclear and cytoplasmic fractions were analyzed for HSF1 by immunoblotting. c-fos and SOD1 serve as nuclear (N) and cytoplasmic (C) markers, respectively. (B) HSF1+/+ MEFs were treated with HSF1A for 6 h or 9 h or heat shocked at 42°C for 2 h in the presence (+) or absence (−) of phosphatase inhibitors (INHIB). (C) HSF1+/+ MEFs were pretreated with 250 µM DTT for 1 h prior to the addition of 80 µM HSF1A for 15 h or a 2-h heat shock at 42°C followed by a 15-h recovery. For comparison purposes, the heat-shocked samples are shown at a lower exposure than the HSF1A-treated samples. (D) HSF1+/+ MEFs were treated with either DMSO or HSF1A (30, 50, or 80 µM) for 15 h at 37°C. For synergistic activation of Hsp70 expression, wild-type MEFs were treated with either DMSO or HSF1A (30, 50, or 80 µM) for 1 h at 37°C prior to a 1-h heat shock at 40°C followed by a 15-h recovery period at 37°C. For control purposes, Hsp70 expression following a 1-h heat shock at 42°C, and a 15-h recovery is shown. SOD1 serves as the protein loading control.
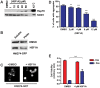
Figure 4. HSF1A suppresses aggregation and cytotoxicity in a cell culture model of Huntington disease.
(A) HD-Q74 PC12 cells were incubated with increasing concentrations of HSF1A for 15 h or heat shocked for 2 h at 42°C, followed by a 15-h recovery and Hsp70 levels analyzed by immunoblotting, with SOD1 as a loading control. (B) PC12 cells were pretreated with either DMSO or 10 µM HSF1A for 15 h and doxycycline added to 1 µg/ml, with further incubation for 48 h. Equal amounts of the soluble and insoluble fractions were analyzed for the presence of httQ74-GFP by immunoblotting with anti-GFP antibody. (C) Fluorescence pattern for httQ74-GFP analyzed microscopically in cells pretreated with DMSO or HSF1A prior to induction of httQ74-GFP expression. (D) Quantification of (C) by counting the number of cells containing aggregates expressed as a percentage of the total number of cells counted. For each treatment, approximately 800 HD-Q74 PC12 cells were evaluated. **p<0.01; ***p<0.001. (E) PC12 cells were pretreated with 4 µM HSF1A for 15 h, before the addition of doxycycline (Dox) to 1 µg/ml followed by a 5-d incubation. Cell viability was assayed by the XTT viability assay. ***p<0.001.
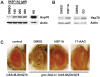
Figure 5. HSF1A promotes Hsp70 expression and reduces polyQ toxicity in fruit flies.
(A) Drosophila S2 cells were treated with increasing concentrations of HSF1A, or heat shocked (HS) at 37°C for 1 h. Hsp70 expression was analyzed by immunoblotting, with actin as a loading control. (B) W1118 flies were raised on food supplemented with DMSO, 5 mM HSF1A, or 0.15 mM geldanamycin (GD) for 3 d. Total protein was extracted from flies and analyzed for Hsp70 expression by immunoblotting as for (A). (C) UAS-MJDtrQ78 flies were crossed to gmr-GAL4 flies in the chronic presence of food supplemented with DMSO, 400 µM HSF1A, or 5 µM 17-AAG. Reductions in eye morphological defects and depigmentation, caused by polyQ-protein expression, are observed with HSF1A and 17-AAG treatment. The lowest effective concentration of HSF1A at which reductions in defects was observed was 400 µM. Control flies are UAS-MJDtrQ78 flies lacking the Gal4 transcription factor. These data are representative of three independent experiments.
Figure 6. HSF1A is unlikely to be an inhibitor of Hsp90.
(A) Yeast cells were treated with either 10 µM HSF1A, 10 µM geldanamycin, or 10 µM radicicol and growth was assessed as in Figure 1C. (B) Purified Hsp90α was incubated with increasing concentrations of 17-AAG or HSF1A for 30 min at 4°C and then incubated with 1 µM geldanamycin-biotin for 1 h at 4°C. (C) Geldanamycin-biotin–bound Hsp90 was captured using neutravidin-agarose beads and analyzed by immunoblot analysis. Hsp90α was incubated with either 10 µM geldanamycin-biotin (GD-B) or 100 µM HSF1A-biotin (HSF1A-B) and was analyzed as described in Figure 2B.
Figure 7. HSF1A-biotin associates with the TRiC/CCT complex.
(A) Cell extracts from mouse embryonic fibroblasts were incubated with 100 µM HSF1A-biotin, and associated proteins were then purified with neutravidin-agarose beads. HSF1A-biotin-interacting proteins were resolved by SDS-PAGE and visualized by silver staining. (B) HSF1A-biotin-interacting proteins were purified as in (A) from HeLa cell extracts and analyzed by immunoblot analysis for Tcp1, Cct8, and Hsp90. (C) HSF1A-biotin-interacting proteins were purified as in (A) from yeast cell extracts and analyzed by immunoblot analysis for yTcp1.
Similar articles
- Heat shock transcription factor 1-activating compounds suppress polyglutamine-induced neurodegeneration through induction of multiple molecular chaperones.
Fujikake N, Nagai Y, Popiel HA, Okamoto Y, Yamaguchi M, Toda T. Fujikake N, et al. J Biol Chem. 2008 Sep 19;283(38):26188-97. doi: 10.1074/jbc.M710521200. Epub 2008 Jul 16. J Biol Chem. 2008. PMID: 18632670 Free PMC article. - Hsf1 on a leash - controlling the heat shock response by chaperone titration.
Masser AE, Ciccarelli M, Andréasson C. Masser AE, et al. Exp Cell Res. 2020 Nov 1;396(1):112246. doi: 10.1016/j.yexcr.2020.112246. Epub 2020 Aug 27. Exp Cell Res. 2020. PMID: 32861670 Review. - Protein phosphatase 5 is a negative modulator of heat shock factor 1.
Conde R, Xavier J, McLoughlin C, Chinkers M, Ovsenek N. Conde R, et al. J Biol Chem. 2005 Aug 12;280(32):28989-96. doi: 10.1074/jbc.M503594200. Epub 2005 Jun 20. J Biol Chem. 2005. PMID: 15967796 - Heat shock factor 1 ameliorates proteotoxicity in cooperation with the transcription factor NFAT.
Hayashida N, Fujimoto M, Tan K, Prakasam R, Shinkawa T, Li L, Ichikawa H, Takii R, Nakai A. Hayashida N, et al. EMBO J. 2010 Oct 20;29(20):3459-69. doi: 10.1038/emboj.2010.225. Epub 2010 Sep 10. EMBO J. 2010. PMID: 20834230 Free PMC article. - [Molecular therapy targeting protein misfolding and aggregation for the polyglutamine diseases].
Nagai Y. Nagai Y. Rinsho Shinkeigaku. 2009 Nov;49(11):913-6. doi: 10.5692/clinicalneurol.49.913. Rinsho Shinkeigaku. 2009. PMID: 20030247 Review. Japanese.
Cited by
- The biology of proteostasis in aging and disease.
Labbadia J, Morimoto RI. Labbadia J, et al. Annu Rev Biochem. 2015;84:435-64. doi: 10.1146/annurev-biochem-060614-033955. Epub 2015 Mar 12. Annu Rev Biochem. 2015. PMID: 25784053 Free PMC article. Review. - The yeast Hsp70 Ssa1 is a sensor for activation of the heat shock response by thiol-reactive compounds.
Wang Y, Gibney PA, West JD, Morano KA. Wang Y, et al. Mol Biol Cell. 2012 Sep;23(17):3290-8. doi: 10.1091/mbc.E12-06-0447. Epub 2012 Jul 18. Mol Biol Cell. 2012. PMID: 22809627 Free PMC article. - Therapeutic Approaches for Inhibition of Protein Aggregation in Huntington's Disease.
Kim S, Kim KT. Kim S, et al. Exp Neurobiol. 2014 Mar;23(1):36-44. doi: 10.5607/en.2014.23.1.36. Epub 2014 Mar 27. Exp Neurobiol. 2014. PMID: 24737938 Free PMC article. Review. - Protein Quality Control by Molecular Chaperones in Neurodegeneration.
Ciechanover A, Kwon YT. Ciechanover A, et al. Front Neurosci. 2017 Apr 6;11:185. doi: 10.3389/fnins.2017.00185. eCollection 2017. Front Neurosci. 2017. PMID: 28428740 Free PMC article. Review. - Heat shock factor 1 drives regulatory T-cell induction to limit murine intestinal inflammation.
Collins CB, Nguyen TT, Leddy RS, Alula KM, Yeckes AR, Strassheim D, Aherne CM, Luck ME, Karoor V, Jedlicka P, Pierce A, de Zoeten EF. Collins CB, et al. Mucosal Immunol. 2024 Feb;17(1):94-110. doi: 10.1016/j.mucimm.2023.11.003. Epub 2023 Nov 7. Mucosal Immunol. 2024. PMID: 37944754 Free PMC article.
References
- Chiti F, Dobson C. M. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. - PubMed
- Orr H. T, Zoghbi H. Y. Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575–621. - PubMed
- Caughey B, Lansbury P. T. Protofibrils, pores, fibrils, and neurodegeneration: separating the responsible protein aggregates from the innocent bystanders. Annu Rev Neurosci. 2003;26:267–298. - PubMed
- Young J. C, Agashe V. R, Siegers K, Hartl F. U. Pathways of chaperone-mediated protein folding in the cytosol. Nat Rev Mol Cell Biol. 2004;5:781–791. - PubMed
- Fujimoto M, Takaki E, Hayashi T, Kitaura Y, Tanaka Y, et al. Active HSF1 significantly suppresses polyglutamine aggregate formation in cellular and mouse models. J Biol Chem. 2005;280:34908–34916. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01-GM059911/GM/NIGMS NIH HHS/United States
- GM076954/GM/NIGMS NIH HHS/United States
- F32 GM076954/GM/NIGMS NIH HHS/United States
- R01 NS065890/NS/NINDS NIH HHS/United States
- R01 GM059911/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical