FACS-based selection of tandem tetracysteine peptides with improved ReAsH brightness in live cells - PubMed (original) (raw)
FACS-based selection of tandem tetracysteine peptides with improved ReAsH brightness in live cells
Schuyler B Van Engelenburg et al. Chembiochem. 2010.
Abstract
Non-perturbing and site-specific in vivo protein labeling methods are highly desired as they allow researchers to probe complex cellular functions. The biarsenical/tetracysteine labeling system allows in situ fluorescent labeling of intracellular proteins which have been appended with small (12 amino acids) genetically encoded peptide tags. In this work we present the in vivo selection of semi-randomized tandem tetracysteine peptides with improved biarsenical (ReAsH) fluorescent brightness (~2-fold) relative to a single tetracysteine motif or rationally designed 3-fold tetracysteine repeat. We found that Fluorescence Activated Cell Sorting by direct ReAsH excitation as opposed to FRET-mediated ReAsH excitation was optimal for selecting 3×Tetracysteine peptides with enhanced brightness. The selected multimer-tetracysteine peptides display enhanced properties due to higher order ReAsH/3×Tetracysteine dye stoichiometries as opposed to enhancement of the individual core tetracysteine photophysical properties. In summary, we have isolated new 3×Tetracysteine motifs with improved ReAsH brightness in live cells. These modular tags should provide enhanced contrast for live cell imaging applications where small tag size (~4.8 KDa) is a requisite for protein labeling.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
Figure 1
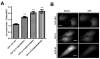
FACS-based selection of GFP-3×TC peptides with improved ReAsH brightness. (A) Library approach for recombination based cloning of GFP-3×TC variants. Core tetracysteine motif (TetCys) indicated in red. (B) FACS analysis showing ReAsH emission as a function of GFP emission for sorts 5 through 8. Histograms were normalized to an isogenic HEK293FT cell line stably expressing GFP-1×TC which was analyzed alongside the 3×TC library for each sort. (C) Selected GFP-3×TC clones display increased ReAsH fluorescence and decreased GFP emission, due to either quenching or decreased GFP-3×TC expression when compared with the core GFP-1×TC and rationally designed GFP-3×TC (GS spacers between each TC motif) proteins. The individual histograms are plotted on the right and top.
Figure 2
Single cell analysis of TC peptides selected for increased ReAsH brightness. (A) Single cell fluorescence microscopy of live HeLa cells reveals enhanced ReAsH fluorescence ratios (~2-fold; ReAsH/GFP) when compared with GFP-1×TC (n ≥ 60 individual cells from at least 3 independent experiments; *** P < 0.0001). Error bars represent SEM. (B) Representative ReAsH fluorescence micrographs of GFP-TC expressing HeLa cells. Images were normalized to the relative GFP expression and rendered at equivalent contrast levels. Scale bars represent 10 μm.
Figure 3
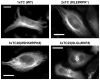
Representative micrographs of ReAsH labeled TC-α-tubulin in HeLa cells. Protein variants 2×TC and 3×TC20 are modular biarsenical binding epitopes which efficiently label the protein α-tubulin. The construct 3×TC22-α-tubulin was unable to form and/or be labeled with ReAsH upon transient expression of this protein suggesting that this 3×TC motif may perturb this proteins function in vivo. Scale bars represent 10 μm.
Figure 4
In vitro titration of selected recombinant GFP-TC peptides demonstrates increased ReAsH:peptide stoichiometries. (A) Fluorescence detected titration of GFP-TC variants shows increased signals with elevated saturation points when compared with the single core GFP-TC motif (n ≥ 3 from three independent experiments for each genotype). (B) Table comparison of the apparent stoichiometries and equilibrium dissociation constants for the selected GFP-3×TC20, -2×TC1, and -1×TC (WT) proteins. Mass spectrometry measurements of ReAsH saturated (~5-fold the number of binding sites) GFP-TC clones are in close agreement with the apparent stoichiometries estimated from the equilibrium titration experiments. It is important to note that under the conditions for ReAsH titration, estimation of the equilibrium dissociation constant is highly inaccurate, but more appropriate for estimation of the stoichiometry of the complex. Error bars represent SD.
Similar articles
- The biarsenical dye Lumio exhibits a reduced ability to specifically detect tetracysteine-containing proteins within live cells.
Hearps AC, Pryor MJ, Kuusisto HV, Rawlinson SM, Piller SC, Jans DA. Hearps AC, et al. J Fluoresc. 2007 Nov;17(6):593-7. doi: 10.1007/s10895-007-0225-x. Epub 2007 Sep 6. J Fluoresc. 2007. PMID: 17805945 - Bipartite tetracysteine display requires site flexibility for ReAsH coordination.
Goodman JL, Fried DB, Schepartz A. Goodman JL, et al. Chembiochem. 2009 Jul 6;10(10):1644-7. doi: 10.1002/cbic.200900207. Chembiochem. 2009. PMID: 19533719 Free PMC article. - ReAsH/FlAsH labeling and image analysis of tetracysteine sensor proteins in cells.
Irtegun S, Ramdzan YM, Mulhern TD, Hatters DM. Irtegun S, et al. J Vis Exp. 2011 Aug 31;(54):2857. doi: 10.3791/2857. J Vis Exp. 2011. PMID: 21897361 Free PMC article. - Flow cytometry of fluorescent proteins.
Telford WG, Hawley T, Subach F, Verkhusha V, Hawley RG. Telford WG, et al. Methods. 2012 Jul;57(3):318-30. doi: 10.1016/j.ymeth.2012.01.003. Epub 2012 Jan 24. Methods. 2012. PMID: 22293036 Review. - Ultrahigh-throughput FACS-based screening for directed enzyme evolution.
Yang G, Withers SG. Yang G, et al. Chembiochem. 2009 Nov 23;10(17):2704-15. doi: 10.1002/cbic.200900384. Chembiochem. 2009. PMID: 19780076 Review.
Cited by
- Visualization of Spirochetes by Labeling Membrane Proteins With Fluorescent Biarsenical Dyes.
Hillman C, Stewart PE, Strnad M, Stone H, Starr T, Carmody A, Evans TJ, Carracoi V, Wachter J, Rosa PA. Hillman C, et al. Front Cell Infect Microbiol. 2019 Aug 20;9:287. doi: 10.3389/fcimb.2019.00287. eCollection 2019. Front Cell Infect Microbiol. 2019. PMID: 31482073 Free PMC article. - A Multicolor Split-Fluorescent Protein Approach to Visualize Listeria Protein Secretion in Infection.
Batan D, Braselmann E, Minson M, Nguyen DMT, Cossart P, Palmer AE. Batan D, et al. Biophys J. 2018 Jul 17;115(2):251-262. doi: 10.1016/j.bpj.2018.03.016. Epub 2018 Apr 10. Biophys J. 2018. PMID: 29653838 Free PMC article. - Elucidating virus entry using a tetracysteine-tagged virus.
Mohl BP, Roy P. Mohl BP, et al. Methods. 2017 Aug 15;127:23-29. doi: 10.1016/j.ymeth.2017.08.004. Epub 2017 Aug 9. Methods. 2017. PMID: 28802715 Free PMC article. Review. - Imaging Translational and Post-Translational Gene Regulatory Dynamics in Living Cells with Antibody-Based Probes.
Lyon K, Stasevich TJ. Lyon K, et al. Trends Genet. 2017 May;33(5):322-335. doi: 10.1016/j.tig.2017.02.003. Epub 2017 Mar 27. Trends Genet. 2017. PMID: 28359585 Free PMC article. Review. - Microfluidics-based selection of red-fluorescent proteins with decreased rates of photobleaching.
Dean KM, Lubbeck JL, Davis LM, Regmi CK, Chapagain PP, Gerstman BS, Jimenez R, Palmer AE. Dean KM, et al. Integr Biol (Camb). 2015 Feb;7(2):263-73. doi: 10.1039/c4ib00251b. Integr Biol (Camb). 2015. PMID: 25477249 Free PMC article.
References
- Chen I, Ting AY. Curr Opin Biotechnol. 2005;16:35–40. - PubMed
- Marks KM, Nolan GP. Nat Methods. 2006;3:591–596. - PubMed
- Griffin BA, Adams SR, Tsien RY. Science. 1998;281:269–272. - PubMed
- Hoffmann C, Gaietta G, Bunemann M, Adams SR, Oberdorff-Maass S, Behr B, Vilardaga JP, Tsien RY, Ellisman MH, Lohse MJ. Nat Methods. 2005;2:171–176. - PubMed
- Gaietta G, Deerinck TJ, Adams SR, Bouwer J, Tour O, Laird DW, Sosinsky GE, Tsien RY, Ellisman MH. Science. 2002;19:503–507. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials