Analysis of ribosome biogenesis factor-modules in yeast cells depleted from pre-ribosomes - PubMed (original) (raw)
Analysis of ribosome biogenesis factor-modules in yeast cells depleted from pre-ribosomes
Juliane Merl et al. Nucleic Acids Res. 2010 May.
Abstract
Formation of eukaryotic ribosomes requires more than 150 biogenesis factors which transiently interact with the nascent ribosomal subunits. Previously, many pre-ribosomal intermediates could be distinguished by their protein composition and rRNA precursor (pre-rRNA) content. We purified complexes of ribosome biogenesis factors from yeast cells in which de novo synthesis of rRNA precursors was down-regulated by genetic means. We compared the protein composition of these largely pre-rRNA free assemblies with the one of analogous pre-ribosomal preparations by semi-quantitative mass spectrometry. The experimental setup minimizes the possibility that the analysed pre-rRNA free protein modules were derived from (partially) disrupted pre-ribosomal particles and provides thereby strong evidence for their pre-ribosome independent existence. In support of the validity of this approach (i) the predicted composition of the analysed protein modules was in agreement with previously described rRNA-free complexes and (ii) in most of the cases we could identify new candidate members of reported protein modules. An unexpected outcome of these analyses was that free large ribosomal subunits are associated with a specific set of ribosome biogenesis factors in cells where neo-production of nascent ribosomes was blocked. The data presented strengthen the idea that assembly of eukaryotic pre-ribosomal particles can result from transient association of distinct building blocks.
Figures
Figure 1.
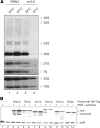
Analysis of cellular pre-rRNA and ribosome biogenesis factor levels after shut down of rRNA de novo synthesis. (A) Northern hybridization analysis of precursor and mature rRNA species of both ribosomal subunits was performed on RNA extracts from whole cells. Yeast strains with RRN3 and with rrn3-8 background were analysed at permissive (24°C) and restrictive (3 h 37°C) temperature. RNA from equal number of cells was loaded and different oligonucleotides (‘Materials and methods’ section) were used for detection of the different indicated (pre-) rRNA species (B) Protein levels of TAP–tagged ribosome biogenesis factors in RRN3 (rRNA synthesis +) and in rrn3-8 background (rRNA synthesis –) were analysed at restrictive (3h, 37°C) temperature by western blotting using PAP visualization reagent. Equal loading was controlled by determination of the protein level of rpS8.
Figure 2.
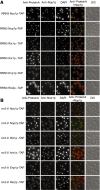
Subcellular localization of TAP–tagged ribosome biogenesis factors after shut down of Pol I transcription. The localization of the indicated tagged proteins in yeast strains with (A) RRN3 and with (B) rrn3-8 background after 3 h shift to 37°C was analysed by fluorescence microscopy using an antibody directed against the Protein A moiety of the TAP-tag. Nucleolar structures were visualized by an anti–Nop1p antibody, yeast nuclei were stained with DAPI. Additionally, the ProteinA-signal (red) and the staining for Nop1p (green) were overlaid for better visualization of subcellular distribution. Whole yeast cells morphology was visualized by differential contrast (DIC). For better comparison the chosen exposition time for detection of the Protein A was not changed during analysis of the different tagged proteins. This results in slightly overexposed pictures for Nop7p-TAP and Arx1p-TAP.
Figure 3.
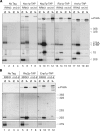
Co-purification of pre-rRNA with different TAP-tagged biogenesis factors of the (A) large and (B) small ribosomal subunit after shut down of Pol-I transcription. Northern hybridization analysis of precursor rRNA species of both ribosomal subunits was performed on RNA extracted from whole cell extracts (IN) and from affinity purified TAP-tagged (A) Noc1p, Nop7p, Rix1p, Arx1p, (B) Rio2p and Enp1p (IP). Yeast strains carrying the RRN3 wild-type allele or the rrn3-8 allele were analysed at restrictive (3 h, 37°C) temperature. Different oligonucleotides (‘Materials and methods’ section) were used for detection of the different indicated (pre-) rRNA species. In parallel the amounts of tagged protein in input and affinity purified fractions were determined by western blotting using PAP visualization reagent. For each precipitation sample same signal intensities in IN and IP lanes reflect a purification recovery of 1%.
Figure 4.
Co-purification of mature 25S and 18S rRNA with TAP-tagged Rix1p and Arx1p after shut down of Pol-I transcription. (A) Northern hybridization analysis of mature rRNA species of both ribosomal subunits was performed on RNA extracted from whole cell extracts (IN) and from affinity purified TAP-tagged Rix1p and Arx1p (IP). Yeast strains carrying the RRN3 wild-type allele or the rrn3-8 allele were analysed at restrictive (3 h, 37°C) temperature. Different oligonucleotides ‘Materials and methods’ section) were used for detection of the indicated 18S and 25S rRNA species. Same signal intensities in IN and IP lanes reflect a purification recovery of 1%. (B) The ratio of affinity precipitation efficiencies for 25S and 18S rRNAs was calculated and normalized to the one found in the untagged wild-type yeast strain using MultiGauge V3.0 (Fujifilm). (C) Sedimentation behaviour of TAP–tagged Arx1p was analysed on sucrose density gradients with cellular extracts of strains carrying the RRN3 wild-type allele (rRNA synthesis +) or the rrn3-8 allele (rRNA synthesis −) after 3 h shift to restrictive temperature. Distribution of ribosomal particles (40S, 60S, 80S, polysomes) in the gradient was determined by OD254 measurement (data not shown) and western blot analysis of the gradient fractions using an anti-rpS8 antibody. The amount of TAP-tagged Arx1p in each fraction and in the input-sample (IN) was also visualized by western blot analysis.
Figure 5.
Schematic view of relative protein quantitation set up. A typical MS spectrum is shown, where each peak represents a mixture of sequence-identical but differentially iTRAQ-labelled peptides from affinity purifications of wild-type and rrn3-8 mutant strains. In the MSMS–mode peptides are selected for fragmentation, yielding in peptide fragments with sequence specific m/z ratios used for identification of the respective protein by database search. In addition the iTRAQ reporter ions of different masses are released and are used for relative quantitation and subsequent determination of pre-rRNA-dependent or pre-rRNA-independent co-purifications.
Figure 6.
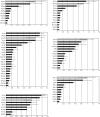
Semi-quantitative comparison of co-purifying ribosome biogenesis factors from cells with or without ongoing rRNA synthesis. The diagrams show the ratio of identified proteins co-purifying with TAP-tagged bait proteins from cells carrying the RRN3 wild-type and the rrn3-8 mutant allele. The bars indicate the average value of the calculated RRN3:rrn3-8 ratios for all identified peptides of the indicated protein. Error bars represent the standard deviation of these ratios (P < 0.05). The ratio of the bait protein (highlighted by a grey bar) is set to one. For co-purified proteins, ratios >0.5 were considered to reflect associations with bait proteins barely influenced by the absence of de novo rRNA synthesis. Association of proteins with intermediate ratios between 0.25 and 0.5 were classified to be stronger affected by the shutoff of rRNA synthesis. Nevertheless they are still significantly co-purified in the rrn3-8 mutant. Associations of proteins with ratios below 0.25 seem to be strongly dependent on rRNA de novo synthesis. Two independent purifications for the indicated bait proteins were performed and for each purification mass spectrometry analysis was repeated once. Quantitation of one representative experiment is shown.
Similar articles
- Disruption of ribosome assembly in yeast blocks cotranscriptional pre-rRNA processing and affects the global hierarchy of ribosome biogenesis.
Talkish J, Biedka S, Jakovljevic J, Zhang J, Tang L, Strahler JR, Andrews PC, Maddock JR, Woolford JL Jr. Talkish J, et al. RNA. 2016 Jun;22(6):852-66. doi: 10.1261/rna.055780.115. Epub 2016 Apr 1. RNA. 2016. PMID: 27036125 Free PMC article. - Interrelationships between yeast ribosomal protein assembly events and transient ribosome biogenesis factors interactions in early pre-ribosomes.
Jakob S, Ohmayer U, Neueder A, Hierlmeier T, Perez-Fernandez J, Hochmuth E, Deutzmann R, Griesenbeck J, Tschochner H, Milkereit P. Jakob S, et al. PLoS One. 2012;7(3):e32552. doi: 10.1371/journal.pone.0032552. Epub 2012 Mar 14. PLoS One. 2012. PMID: 22431976 Free PMC article. - Role of the yeast ribosomal protein L16 in ribosome biogenesis.
Espinar-Marchena FJ, Fernández-Fernández J, Rodríguez-Galán O, Fernández-Pevida A, Babiano R, de la Cruz J. Espinar-Marchena FJ, et al. FEBS J. 2016 Aug;283(16):2968-85. doi: 10.1111/febs.13797. Epub 2016 Jul 15. FEBS J. 2016. PMID: 27374275 - Nuclear export and cytoplasmic maturation of ribosomal subunits.
Zemp I, Kutay U. Zemp I, et al. FEBS Lett. 2007 Jun 19;581(15):2783-93. doi: 10.1016/j.febslet.2007.05.013. Epub 2007 May 11. FEBS Lett. 2007. PMID: 17509569 Review. - Ribosome biogenesis: of knobs and RNA processing.
Granneman S, Baserga SJ. Granneman S, et al. Exp Cell Res. 2004 May 15;296(1):43-50. doi: 10.1016/j.yexcr.2004.03.016. Exp Cell Res. 2004. PMID: 15120992 Review.
Cited by
- Hierarchical recruitment into nascent ribosomes of assembly factors required for 27SB pre-rRNA processing in Saccharomyces cerevisiae.
Talkish J, Zhang J, Jakovljevic J, Horsey EW, Woolford JL Jr. Talkish J, et al. Nucleic Acids Res. 2012 Sep 1;40(17):8646-61. doi: 10.1093/nar/gks609. Epub 2012 Jun 26. Nucleic Acids Res. 2012. PMID: 22735702 Free PMC article. - Studies on the assembly characteristics of large subunit ribosomal proteins in S. cerevisae.
Ohmayer U, Gamalinda M, Sauert M, Ossowski J, Pöll G, Linnemann J, Hierlmeier T, Perez-Fernandez J, Kumcuoglu B, Leger-Silvestre I, Faubladier M, Griesenbeck J, Woolford J, Tschochner H, Milkereit P. Ohmayer U, et al. PLoS One. 2013 Jul 10;8(7):e68412. doi: 10.1371/journal.pone.0068412. Print 2013. PLoS One. 2013. PMID: 23874617 Free PMC article. - Disruption of ribosome assembly in yeast blocks cotranscriptional pre-rRNA processing and affects the global hierarchy of ribosome biogenesis.
Talkish J, Biedka S, Jakovljevic J, Zhang J, Tang L, Strahler JR, Andrews PC, Maddock JR, Woolford JL Jr. Talkish J, et al. RNA. 2016 Jun;22(6):852-66. doi: 10.1261/rna.055780.115. Epub 2016 Apr 1. RNA. 2016. PMID: 27036125 Free PMC article. - Rrp5p, Noc1p and Noc2p form a protein module which is part of early large ribosomal subunit precursors in S. cerevisiae.
Hierlmeier T, Merl J, Sauert M, Perez-Fernandez J, Schultz P, Bruckmann A, Hamperl S, Ohmayer U, Rachel R, Jacob A, Hergert K, Deutzmann R, Griesenbeck J, Hurt E, Milkereit P, Baßler J, Tschochner H. Hierlmeier T, et al. Nucleic Acids Res. 2013 Jan;41(2):1191-210. doi: 10.1093/nar/gks1056. Epub 2012 Dec 2. Nucleic Acids Res. 2013. PMID: 23209026 Free PMC article. - Mechanistic insight into eukaryotic 60S ribosomal subunit biogenesis by cryo-electron microscopy.
Greber BJ. Greber BJ. RNA. 2016 Nov;22(11):1643-1662. doi: 10.1261/rna.057927.116. RNA. 2016. PMID: 27875256 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases