microRNAs in heart disease: putative novel therapeutic targets? - PubMed (original) (raw)
Review
microRNAs in heart disease: putative novel therapeutic targets?
Gianluigi Condorelli et al. Eur Heart J. 2010 Mar.
Abstract
microRNAs (miRs) are short, approximately 22-nucleotide-long non-coding RNAs involved in the control of gene expression. They guide ribonucleoprotein complexes that effect translational repression or messenger RNA degradation to targeted messenger RNAs. miRs were initially thought to be peculiar to the developmental regulation of the nematode worm, in which they were first described in 1993. Since then, hundreds of different miRs have been reported in diverse organisms, and many have been implicated in the regulation of physiological processes of adult animals. Of importance, misexpression of miRs has been uncovered as a pathogenic mechanism in several diseases. Here, we first outline the biogenesis and mechanism of action of miRs, and then discuss their relevance to heart biology, pathology, and medicine.
Figures
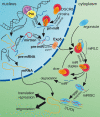
Figure 1
Schematic of microRNA biogenesis and action. See text for explanation. The mature microRNA sequence is given in red. TF, transcription factor; Pol, RNA polymerase II or III; Exp5, exportin 5.
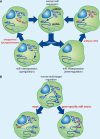
Figure 2
Schematic overview of strategies used to alter microRNA expression. (A) Cells express a microRNA profile that can become altered with disease. Antisense oligonucleotides, such as antagomirs, sponges, and erasers (in red) can capture microRNAs for knockdown or sequestrate inappropriately overexpressed microRNAs, whereas artificially introduced microRNAs (in red) can be used to overexpress microRNAs or, potentially, to replace expression of downregulated ones. These strategies have the potential to affect large numbers of different targets (for simplicity, only one target mRNA per microRNA is represented). (B) Masks and gene-specific microRNA mimics (in red) can be used to affect single targets specifically (mRNAs in different shades of blue represent a set affected by a given microRNA).
Similar articles
- Interactions between microRNAs and long non-coding RNAs in cardiac development and repair.
Rotini A, Martínez-Sarrà E, Pozzo E, Sampaolesi M. Rotini A, et al. Pharmacol Res. 2018 Jan;127:58-66. doi: 10.1016/j.phrs.2017.05.029. Epub 2017 Jun 17. Pharmacol Res. 2018. PMID: 28629929 Review. - MicroRNAs differentially regulated in cardiac and skeletal muscle in health and disease: potential drug targets?
Winbanks CE, Ooi JY, Nguyen SS, McMullen JR, Bernardo BC. Winbanks CE, et al. Clin Exp Pharmacol Physiol. 2014 Sep;41(9):727-37. doi: 10.1111/1440-1681.12281. Clin Exp Pharmacol Physiol. 2014. PMID: 25115402 Review. - MicroRNAs in skeletal and cardiac muscle development.
Callis TE, Chen JF, Wang DZ. Callis TE, et al. DNA Cell Biol. 2007 Apr;26(4):219-25. doi: 10.1089/dna.2006.0556. DNA Cell Biol. 2007. PMID: 17465888 Review. - MicroRNAs challenge the status quo of therapeutic targeting.
Sayed D, Rane S, Abdellatif M. Sayed D, et al. J Cardiovasc Transl Res. 2009 Mar;2(1):100-7. doi: 10.1007/s12265-008-9052-y. Epub 2008 Sep 9. J Cardiovasc Transl Res. 2009. PMID: 20559973 Review. - miRNAs got rhythm.
Elton TS, Martin MM, Sansom SE, Belevych AE, Györke S, Terentyev D. Elton TS, et al. Life Sci. 2011 Feb 28;88(9-10):373-83. doi: 10.1016/j.lfs.2010.11.022. Epub 2010 Dec 3. Life Sci. 2011. PMID: 21130781 Review.
Cited by
- Molecular biomarkers in diabetes mellitus (DM).
Aghaei Zarch SM, Dehghan Tezerjani M, Talebi M, Vahidi Mehrjardi MY. Aghaei Zarch SM, et al. Med J Islam Repub Iran. 2020 Apr 1;34:28. doi: 10.34171/mjiri.34.28. eCollection 2020. Med J Islam Repub Iran. 2020. PMID: 32617267 Free PMC article. Review. - Micrornas and Cardiovascular Diseases: From Bench to Bedside.
De Rosa R, Polito MV, Benvenga R, De Angelis E, Piscione F, Galasso G. De Rosa R, et al. Transl Med UniSa. 2018 Mar 31;17:12-18. eCollection 2017 Jul. Transl Med UniSa. 2018. PMID: 30050875 Free PMC article. - Novel concepts in HDL pharmacology.
Remaley AT, Norata GD, Catapano AL. Remaley AT, et al. Cardiovasc Res. 2014 Aug 1;103(3):423-8. doi: 10.1093/cvr/cvu141. Epub 2014 Jun 20. Cardiovasc Res. 2014. PMID: 24951539 Free PMC article. Review. - LIPCAR: a mitochondrial lnc in the noncoding RNA chain?
Dorn GW 2nd. Dorn GW 2nd. Circ Res. 2014 May 9;114(10):1548-50. doi: 10.1161/CIRCRESAHA.114.304028. Circ Res. 2014. PMID: 24812345 Free PMC article. No abstract available. - Role of microRNA in diabetic cardiomyopathy: From mechanism to intervention.
Guo R, Nair S. Guo R, et al. Biochim Biophys Acta Mol Basis Dis. 2017 Aug;1863(8):2070-2077. doi: 10.1016/j.bbadis.2017.03.013. Epub 2017 Mar 24. Biochim Biophys Acta Mol Basis Dis. 2017. PMID: 28344129 Free PMC article. Review.
References
- Carninci P, Yasuda J, Hayashizaki Y. Multifaceted mammalian transcriptome. Curr Opin Cell Biol. 2008;3:274–280. - PubMed
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. - PubMed
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. - PubMed
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed