ATM deficiency sensitizes mantle cell lymphoma cells to poly(ADP-ribose) polymerase-1 inhibitors - PubMed (original) (raw)
ATM deficiency sensitizes mantle cell lymphoma cells to poly(ADP-ribose) polymerase-1 inhibitors
Chris T Williamson et al. Mol Cancer Ther. 2010 Feb.
Abstract
Poly(ADP-ribose) polymerase-1 (PARP-1) inhibition is toxic to cells with mutations in the breast and ovarian cancer susceptibility genes BRCA1 or BRCA2, a concept termed synthetic lethality. However, whether this approach is applicable to other human cancers with defects in other DNA repair genes has yet to be determined. The ataxia telangiectasia mutated (ATM) gene is altered in several human cancers including mantle cell lymphoma (MCL). Here, we characterize a panel of MCL cell lines for ATM status and function and investigate the potential for synthetic lethality in MCL in the presence of small-molecule inhibitors of PARP-1. We show that Granta-519 and UPN2 cells have low levels of ATM protein, are defective in DNA damage-induced ATM-dependent signaling, are radiation sensitive, and have cell cycle checkpoint defects: all characteristics of defective ATM function. Significantly, Granta-519 and UPN2 cells were more sensitive to PARP-1 inhibition than were the ATM-proficient MCL cell lines examined. Furthermore, the PARP-1 inhibitor olaparib (known previously as AZD2281/KU-0059436) significantly decreased tumor growth and increased overall survival in mice bearing s.c. xenografts of ATM-deficient Granta-519 cells while producing only a modest effect on overall survival of mice bearing xenografts of the ATM-proficient cell line, Z138. Thus, PARP inhibitors have therapeutic potential in the treatment of MCL, and the concept of synthetic lethality extends to human cancers with ATM alterations.
Figures
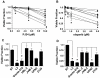
Figure 1. Deficiency of ATM level and function in Granta-519 and UPN2 cell lines
A) Relative levels of expression of ATM, DNA-PKcs, SMC-1 and PARP-1 proteins in the MCL cell lines HBL-2, JVM-2, Granta-519, UPN1, UPN2, MAVER-1 and Z138 compared to a control lymphoblastoid cell line C35ABR (BT) and an A-T patient-derived lymphoblastoid cell line (L3). B) ATM protein levels in panel A were quantitated and normalized to the levels of DNA-PKcs and SMC-1 in BT cells. C) BT, L3 and MCL cell lines were either unirradiated or irradiated with 2 Gy IR and cellular viability was determined after 96 hours using the WST-1 assay. The fraction of viable cells normalized to the untreated control of each cell line is shown. D) Cells were either untreated (black bars) or irradiated with 2 Gy IR, collected either 1 hour (grey bars) or 24 hours (white bars) later and assayed for phospho-Ser-20 histone H3 phosphorylation as described in Materials and Methods. The percentage of cells in mitosis was normalized to the untreated control of each cell line to give the mitotic fraction.
Figure 2. ATM-dependent signaling is reduced in Granta-519 and UPN2 cell lines
MCL cell lines were exposed to 2 Gy IR and harvested following the indicated incubation times. The ATM-proficient lymphoblastoid cell line, BT, is shown in panels C and D as a positive control for ATM-dependent signaling. Whole cell extracts (50 μg total protein) were analyzed by SDS PAGE and immunoblotted for autophosphorylation of ATM on Ser-1981 (P-S1981), phosphorylation of SMC-1 on Ser-957 and Ser-966 (P-S957 and P-S966, respectively), and phosphorylation of KAP1 on Ser-824 (P-S824), as indicated. Total ATM, SMC-1 and KAP1 protein levels are also shown. Actin and DNA-PKcs are shown as loading controls. Panel A, Z138; panel B; UPN1; panel C, UPN2 and panel D, Granta-519. Signaling pathways in BT, L3, HBL-2, JVM-2 and MAVER-1 cells are shown in Supplementary Figures 1 and 2, respectively.
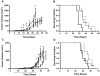
Figure 3. PARP-1 inhibitors preferentially target ATM-deficient MCL cells
BT, L3 and MCL cell lines were exposed to the indicated concentrations of A) PJ34, or B) olaparib or vehicle control for 96 hours and cellular viability was determined by trypan blue exclusion. Cell viability was normalized to the vehicle-treated control for each cell line. Granta-519 (white triangles), HBL-2 (black triangles), JVM-2 (inverted black triangles), MAVER-1 (black diamonds), UPN1 (black circles), UPN2 (white squares), Z138 (black squares), BT (black hexagons), and L3 (white diamonds). Error bars represent the standard error of the mean (SEM). Points marked by an asterisk (*) indicate statistical significance (p<0.05) between ATM-proficient and ATM-deficient MCL cell lines. In panel A the point marked by a cross (‡) indicates a statistically significant difference (p<0.05) between Granta-519 and ATM-proficient MCL cell lines. C) BT, L3 and MCL cell lines were incubated with vehicle alone or PJ34 (10 μM) for 96 hours after which point cell viability was determined using the WST-1 assay. Cell viability was normalized to the vehicle-treated control for each cell line. D) Cells were treated with vehicle alone or olaparib (5 μM). After 96 hours, cell viability was determined as in panel C. Error bars represent the standard error of the mean (SEM). For panels C and D, points marked by an asterisk (*) indicate statistical significance (p<0.05) between BT and L3, and ATM-proficient and ATM-deficient MCL cell lines. The line indicates which experimental parameters are being compared. In each experiment BT cells are compare to L3 cells, while all the MCL cell lines are compared to each other.
Figure 4. Inhibition of PARP-1 is cytotoxic in MCL cells with reduced ATM protein expression
A) Z138 cells were stably transfected with a vector expressing shRNA targeting ATM (ZC-shATM) or GFP (ZC-shGFP) as a negative control as described in Materials and Methods. 50 μg of whole cell extract were run on SDS PAGE and immunoblots were probed for ATM, DNA-PKcs, SMC-1, PARP-1, cyclin D1 and actin protein expression as indicated. B) Z138, ZC-shGFP, or ZC-shATM cells were either unirradiated (-) or irradiated (2 Gy) and harvested following a two hour incubation. Whole cell extracts were probed for phosphorylated ATM (P-S1981), total ATM and DNA-PKcs as indicated. C) Cells were incubated with various concentrations of olaparib and after 96 hours cell viability was determined by trypan blue exclusion. Z138 (black squares), ZC-shGFP (black circles), ZC-shATM (white squares), BT (black diamonds) and L3 (white circles). Points marked by an asterisk (*) indicate statistical significance (p<0.05) between ZC-shATM and Z138/ZC-shGFP. D) BT, L3, Z138, ZC-shGFP and ZC-shATM cells were exposed to olaparib (5 μM) for 96 hours then cellular viability was determining using the WST-1 assay. For each panel, error bars represent the SEM. In each experiment BT cells are compare to L3 cells, while all the MCL cell lines are compared to each other.
Figure 5. Olaparib induces serine-1981 phosphorylation in ATM-proficient MCL cells and apoptosis in ATM-deficient MCL cells
A) Z138 and Granta-519 cells were exposed to olaparib (2.5 μM) or vehicle (VEH, DMSO) for 24, 48, 72 or 96 hours as indicated. Whole cell extracts (50 μg total protein) were run on SDS PAGE, immunoblotted and probed for ATM autophosphorylation at Ser-1981 (P-S1981), total ATM and total SMC-1 as indicated. Quantitation of olaparib-induced P-S1981 ATM compared to total ATM for Z138 is shown below the blot. As a positive control for P-Ser-1981, in the Granta-519 cells, BT cells were irradiated 2 Gy and harvested after 1 hour. B) Cells were treated with 2.5 μM olaparib for 24, 48, 72 or 96 hours as indicated and the proportion of cells undergoing apoptosis was determined using the TUNEL assay. The fraction of TUNEL positive cells was normalized to the untreated sample for each cell line. Results are presented as the percentage of TUNEL positive cells at each time point. Z138 (white bars), Granta-519 (black bars), UPN2 (hatched bars) and UPN1 (grey bars). C) Cells were treated with olaparib as in panel C then assayed for Annexin V and PI staining. The percentage of apoptotic cells (+ for Annexin V and – for PI) is shown. Z138 (white bars), Granta-519 (black bars), UPN2 (hatched bars) and UPN1 (grey bars) as in panel C. D) A model for the mechanism of PARP-1-induced cell death. DNA single strand breaks (SSBs) generated in cells by reactive oxygen species (ROS) or as intermediates during base excision repair (BER) are recognized by PARP-1 and repaired by the base excision and/or DNA single strand break (SSB) repair pathways. Small molecule inhibitors of PARP-1 (for example PJ34 or olaparib) block SSB repair, permitting the conversion of SSBs into DNA double strand breaks (DSBs) during DNA replication. MCL cells with wild-type ATM initiate an appropriate DSB damage response and survive, whereas cells in which ATM function is disrupted have reduced ability to respond to DNA DSBs, resulting in cell death via apoptosis.
Figure 6. Olaparib reduces tumour growth and prolongs survival of mice bearing ATM-deficient xenografts
A) Mice were injected subcutaneously with Granta-519 (ATM-deficient) cells as described in Materials and Methods. Five days later mice were injected intraperitoneally with vehicle alone (circles, solid lines), 25 mg/kg olaparib (squares, dashed lines), or 50 mg/kg olaparib (triangles, dotted lines). Injections of drug/vehicle were continued for 28 consecutive days (indicated by solid line beneath the x axis). Tumour volume was determined as described in Materials and Methods. N = 8 mice for the 0 and 50 mg/kg groups and 9 for the 25 mg/kg group. Error bars represent the SEM. Statistical significance (p > 0.05), as determined by the student T-test between control and olaparib-treated mice is marked either by an asterisk (50 mg/kg group) or an octothorp/number sign (25 mg/kg group). B) Survival curves for the experiment shown in panel A. Solid lines represent mice injected with vehicle alone, dashed lines and dotted lines represent mice treated with 25 mg/kg or 50 mg/kg olaparib, respectively. End point survival at 25 and 50 mg/kg were considered statistically significant (p = 0.0018 and 0.0012, respectively) compared to vehicle treated animals using the Mantel-Cox test. Mean survival times were 28 days (vehicle alone), 35 days (25 mg/kg olaparib) and 40 days for 50 mg/kg olaparib. C) Tumour volume for mice injected with Z138 cells (ATM-proficient) followed by injection with vehicle alone (circles, solid lines), 25 mg/kg olaparib (squares, dashed lines) or 50 mg/kg olaparib (triangles, dotted lines) as described in panel A. N = 10 mice for the 0 and 25 mg/kg groups and 9 for the 50 mg/kg group. Error bars represent the SEM. Points marked by an asterisk (50 mg/kg group) are considered statistically significant (p > 0.05) compared to vehicle-treated controls as determined by the student T-test. D) Survival curves for the experiment shown in panel C. Vehicle alone (solid lines), 25 mg/kg olaparib (dashed lines), 50 mg/kg olaparib (dotted lines). End point survival between 0 and 25 mg/kg were not considered statistically significant (p = 0.316). End point survival between and 0 and 50 mg/kg were considered statistically significant (p = 0.0057) using the Mantel-Cox test. The mean survival of mice receiving no olaparib was 45 days, compared with 45 and 56 days for mice receiving 25 or 50 mg/kg olaparib, respectively.
Similar articles
- Enhanced cytotoxicity of PARP inhibition in mantle cell lymphoma harbouring mutations in both ATM and p53.
Williamson CT, Kubota E, Hamill JD, Klimowicz A, Ye R, Muzik H, Dean M, Tu L, Gilley D, Magliocco AM, McKay BC, Bebb DG, Lees-Miller SP. Williamson CT, et al. EMBO Mol Med. 2012 Jun;4(6):515-27. doi: 10.1002/emmm.201200229. Epub 2012 Mar 13. EMBO Mol Med. 2012. PMID: 22416035 Free PMC article. - The PARP inhibitor olaparib induces significant killing of ATM-deficient lymphoid tumor cells in vitro and in vivo.
Weston VJ, Oldreive CE, Skowronska A, Oscier DG, Pratt G, Dyer MJ, Smith G, Powell JE, Rudzki Z, Kearns P, Moss PA, Taylor AM, Stankovic T. Weston VJ, et al. Blood. 2010 Nov 25;116(22):4578-87. doi: 10.1182/blood-2010-01-265769. Epub 2010 Aug 25. Blood. 2010. PMID: 20739657 - Inhibition of poly(ADP-ribose) polymerase (PARP) and ataxia telangiectasia mutated (ATM) on the chemosensitivity of mantle cell lymphoma to agents that induce DNA strand breaks.
Golla RM, Li M, Shen Y, Ji M, Yan Y, Fu K, Greiner TC, McKeithan TW, Chan WC. Golla RM, et al. Hematol Oncol. 2012 Dec;30(4):175-9. doi: 10.1002/hon.1020. Epub 2011 Dec 14. Hematol Oncol. 2012. PMID: 22170260 - [From poly(ADP-ribose) discovery to PARP inhibitors in cancer therapy].
Schreiber V, Illuzzi G, Héberlé E, Dantzer F. Schreiber V, et al. Bull Cancer. 2015 Oct;102(10):863-73. doi: 10.1016/j.bulcan.2015.07.012. Epub 2015 Sep 15. Bull Cancer. 2015. PMID: 26384693 Review. French. - DNA damage response pathways in tumor suppression and cancer treatment.
Liang Y, Lin SY, Brunicardi FC, Goss J, Li K. Liang Y, et al. World J Surg. 2009 Apr;33(4):661-6. doi: 10.1007/s00268-008-9840-1. World J Surg. 2009. PMID: 19034564 Review.
Cited by
- Strategies to improve radiotherapy with targeted drugs.
Begg AC, Stewart FA, Vens C. Begg AC, et al. Nat Rev Cancer. 2011 Apr;11(4):239-53. doi: 10.1038/nrc3007. Nat Rev Cancer. 2011. PMID: 21430696 Review. - Therapeutic potential of the poly(ADP-ribose) polymerase inhibitor rucaparib for the treatment of sporadic human ovarian cancer.
Ihnen M, zu Eulenburg C, Kolarova T, Qi JW, Manivong K, Chalukya M, Dering J, Anderson L, Ginther C, Meuter A, Winterhoff B, Jones S, Velculescu VE, Venkatesan N, Rong HM, Dandekar S, Udar N, Jänicke F, Los G, Slamon DJ, Konecny GE. Ihnen M, et al. Mol Cancer Ther. 2013 Jun;12(6):1002-15. doi: 10.1158/1535-7163.MCT-12-0813. Epub 2013 May 31. Mol Cancer Ther. 2013. PMID: 23729402 Free PMC article. - Acceleration or Brakes: Which Is Rational for Cell Cycle-Targeting Neuroblastoma Therapy?
Ando K, Nakagawara A. Ando K, et al. Biomolecules. 2021 May 18;11(5):750. doi: 10.3390/biom11050750. Biomolecules. 2021. PMID: 34069817 Free PMC article. Review. - The Elephant and the Blind Men: Making Sense of PARP Inhibitors in Homologous Recombination Deficient Tumor Cells.
De Lorenzo SB, Patel AG, Hurley RM, Kaufmann SH. De Lorenzo SB, et al. Front Oncol. 2013 Sep 11;3:228. doi: 10.3389/fonc.2013.00228. Front Oncol. 2013. PMID: 24062981 Free PMC article. Review. - Tumor protein D52 represents a negative regulator of ATM protein levels.
Chen Y, Kamili A, Hardy JR, Groblewski GE, Khanna KK, Byrne JA. Chen Y, et al. Cell Cycle. 2013 Sep 15;12(18):3083-97. doi: 10.4161/cc.26146. Epub 2013 Aug 21. Cell Cycle. 2013. PMID: 23974097 Free PMC article.
References
- Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–74. - PubMed
- O’Connor MJ, Martin NM, Smith GC. Targeted cancer therapies based on the inhibition of DNA strand break repair. Oncogene. 2007;26:7816–24. - PubMed
- Martin SA, Lord CJ, Ashworth A. DNA repair deficiency as a therapeutic target in cancer. Curr Opin Genet Dev. 2008;18:80–6. - PubMed
- Lord CJ, Ashworth A. Targeted therapy for cancer using PARP inhibitors. Curr Opin Pharmacol. 2008;8:363–9. - PubMed
- Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer. 2008;8:193–204. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous