Persistent donor cell gene expression among human induced pluripotent stem cells contributes to differences with human embryonic stem cells - PubMed (original) (raw)
Persistent donor cell gene expression among human induced pluripotent stem cells contributes to differences with human embryonic stem cells
Zhumur Ghosh et al. PLoS One. 2010.
Abstract
Human induced pluripotent stem cells (hiPSCs) generated by de-differentiation of adult somatic cells offer potential solutions for the ethical issues surrounding human embryonic stem cells (hESCs), as well as their immunologic rejection after cellular transplantation. However, although hiPSCs have been described as "embryonic stem cell-like", these cells have a distinct gene expression pattern compared to hESCs, making incomplete reprogramming a potential pitfall. It is unclear to what degree the difference in tissue of origin may contribute to these gene expression differences. To answer these important questions, a careful transcriptional profiling analysis is necessary to investigate the exact reprogramming state of hiPSCs, as well as analysis of the impression, if any, of the tissue of origin on the resulting hiPSCs. In this study, we compare the gene profiles of hiPSCs derived from fetal fibroblasts, neonatal fibroblasts, adipose stem cells, and keratinocytes to their corresponding donor cells and hESCs. Our analysis elucidates the overall degree of reprogramming within each hiPSC line, as well as the "distance" between each hiPSC line and its donor cell. We further identify genes that have a similar mode of regulation in hiPSCs and their corresponding donor cells compared to hESCs, allowing us to specify core sets of donor genes that continue to be expressed in each hiPSC line. We report that residual gene expression of the donor cell type contributes significantly to the differences among hiPSCs and hESCs, and adds to the incompleteness in reprogramming. Specifically, our analysis reveals that fetal fibroblast-derived hiPSCs are closer to hESCs, followed by adipose, neonatal fibroblast, and keratinocyte-derived hiPSCs.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
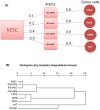
Figure 1. Matrix showing the number of differentially expressed genes P<0.05 and fold-change ≥2 across hESCs, hiPSCs, and donor cell lines.
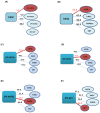
Figure 2. Distance between hiPSC, hESC and donor cells.
(A) Relative distances of the hiPSC states from the corresponding somatic states (donor cells), and from the hESC state. (B) Global clustering among hESC, hiPSC, and donor cells.
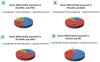
Figure 3. Percentage of differentially expressed genes defines the degree of dissimilarity between hESCs and hiPSCs, hESCs with the donor cell types, and hiPSCs with the 4 different donor cell types.
(A) The distance between hESCs and hiPSCs shows iPS-hFFib to be closest to hESCs. (B) The distance between hESCs and donor cell types shows hFFib to be closest to hESCs. The distance between hiPSCs and 4 different donor cell types shows (C) iPS-hFFib closest to hFFib; (D) iPS-hASC closest to hASC; (E) iPS-hNFib closest to hNFib; and (F) iPS-hKT closest to hKT. (Closest grouping is marked with a red circle).
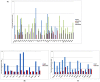
Figure 4. Modes of regulation of the differentially expressed genes across different hiPSCs and their corresponding donor cells compared to hESCs.
77% of the genes have similar expression pattern in iPS-hFFib and hFFib (both upregulated and downreglated). 84% of the genes have similar expression pattern in iPS-hASC and hASC. 85% of the genes have similar expression pattern in iPS-hNFib and hNFib. 96% of the genes have similar expression pattern in iPS-hKT and hKT.
Figure 5. Hierarchical clustering of upregulated genes in the hiPSC and donor cells.
(A) iPS-hFFib and hFFib. (B) iPS-hASC and hASC. (C) iPS-hNFib and hNFib. (D) iPS-hKT and hKT. Hierarchical clustering of the upregulated gene expression data shows that hiPSCs cluster more closely to their corresponding donor cells. This demonstrates that reprogrammed hiPSCs exhibit persistent gene expression from their corresponding donor cells.
Figure 6. Residual signatures of the donor cell specific genes (upregulated in both hiPSCs and donor cells compared to hESC) in hiPSCs.
(A) Expression fold-change of fibroblast specific genes in iPS-hFFib, hFFib, iPS-hNFib, and hNFib. (B) Expression fold-change of adipose cell specific genes in iPS-hASC and hASC. (C) Expression fold-change of keratinocyte specific genes in iPS-hKT and hKT.
Figure 7. Residual signatures of the genes (downregulated in both hiPSCs and donor cells compared to hESC) involved in human embryonic stem cell pluripotency.
(A) Expression fold-change in iPS-hFFib and hFFib. (B) Expression fold-change in iPS-hASC and hASC. (C) Expression fold-change in iPS-hNFib and hNFib. (D) Expression fold-change in iPS-hKT and hKT.
Similar articles
- Comparative study of human-induced pluripotent stem cells derived from bone marrow cells, hair keratinocytes, and skin fibroblasts.
Streckfuss-Bömeke K, Wolf F, Azizian A, Stauske M, Tiburcy M, Wagner S, Hübscher D, Dressel R, Chen S, Jende J, Wulf G, Lorenz V, Schön MP, Maier LS, Zimmermann WH, Hasenfuss G, Guan K. Streckfuss-Bömeke K, et al. Eur Heart J. 2013 Sep;34(33):2618-29. doi: 10.1093/eurheartj/ehs203. Epub 2012 Jul 12. Eur Heart J. 2013. PMID: 22798560 - The Aberrant DNA Methylation Profile of Human Induced Pluripotent Stem Cells Is Connected to the Reprogramming Process and Is Normalized During In Vitro Culture.
Tesarova L, Simara P, Stejskal S, Koutna I. Tesarova L, et al. PLoS One. 2016 Jun 23;11(6):e0157974. doi: 10.1371/journal.pone.0157974. eCollection 2016. PLoS One. 2016. PMID: 27336948 Free PMC article. - Gene and MicroRNA Profiling of Human Induced Pluripotent Stem Cell-Derived Endothelial Cells.
Wang L, Su W, Du W, Xu Y, Wang L, Kong D, Han Z, Zheng G, Li Z. Wang L, et al. Stem Cell Rev Rep. 2015 Apr;11(2):219-27. doi: 10.1007/s12015-014-9582-4. Stem Cell Rev Rep. 2015. PMID: 25618294 - Who Will Win: Induced Pluripotent Stem Cells Versus Embryonic Stem Cells for β Cell Replacement and Diabetes Disease Modeling?
Jacobson EF, Tzanakakis ES. Jacobson EF, et al. Curr Diab Rep. 2018 Oct 20;18(12):133. doi: 10.1007/s11892-018-1109-y. Curr Diab Rep. 2018. PMID: 30343423 Review. - Reprogramming of Keratinocytes as Donor or Target Cells Holds Great Promise for Cell Therapy and Regenerative Medicine.
Zhang Y, Hu W, Ma K, Zhang C, Fu X. Zhang Y, et al. Stem Cell Rev Rep. 2019 Oct;15(5):680-689. doi: 10.1007/s12015-019-09900-8. Stem Cell Rev Rep. 2019. PMID: 31197578 Review.
Cited by
- Does transcription factor induced pluripotency accurately mimic embryo derived pluripotency?
Lowry WE. Lowry WE. Curr Opin Genet Dev. 2012 Oct;22(5):429-34. doi: 10.1016/j.gde.2012.07.003. Epub 2012 Oct 15. Curr Opin Genet Dev. 2012. PMID: 23079387 Free PMC article. Review. - Passage number affects differentiation of sensory neurons from human induced pluripotent stem cells.
Cantor EL, Shen F, Jiang G, Tan Z, Cunningham GM, Wu X, Philips S, Schneider BP. Cantor EL, et al. Sci Rep. 2022 Sep 23;12(1):15869. doi: 10.1038/s41598-022-19018-6. Sci Rep. 2022. PMID: 36151116 Free PMC article. - A circular RNA map for human induced pluripotent stem cells of foetal origin.
Barilani M, Cherubini A, Peli V, Polveraccio F, Bollati V, Guffanti F, Del Gobbo A, Lavazza C, Giovanelli S, Elvassore N, Lazzari L. Barilani M, et al. EBioMedicine. 2020 Jul;57:102848. doi: 10.1016/j.ebiom.2020.102848. Epub 2020 Jun 20. EBioMedicine. 2020. PMID: 32574961 Free PMC article. - Analysis of human and mouse reprogramming of somatic cells to induced pluripotent stem cells. What is in the plate?
Boué S, Paramonov I, Barrero MJ, Izpisúa Belmonte JC. Boué S, et al. PLoS One. 2010 Sep 17;5(9):e12664. doi: 10.1371/journal.pone.0012664. PLoS One. 2010. PMID: 20862250 Free PMC article. - Stem Cell Therapies in Kidney Diseases: Progress and Challenges.
Rota C, Morigi M, Imberti B. Rota C, et al. Int J Mol Sci. 2019 Jun 7;20(11):2790. doi: 10.3390/ijms20112790. Int J Mol Sci. 2019. PMID: 31181604 Free PMC article. Review.
References
- Stojkovic M, Lako M, Strachan T, Murdoch A. Derivation, growth and applications of human embryonic stem cells. Reproduction. 2004;128:259–267. - PubMed
- Nishikawa S-i, Goldstein R, Nierras C. The promise of human induced pluripotent stem cells for research and therapy. Nature Reviews Molecular Cell Biology. 2008;9:725–729. - PubMed
- Sipp D. Gold standards in the diamond age: the commodification of pluripotency. Cell Stem Cell. 2009;5:360–363. - PubMed
Publication types
MeSH terms
Grants and funding
- R01 AI085575-01/AI/NIAID NIH HHS/United States
- R01 AI085575/AI/NIAID NIH HHS/United States
- R01AI085575/AI/NIAID NIH HHS/United States
- DP2 OD004437/OD/NIH HHS/United States
- DP2 OD004437-01/OD/NIH HHS/United States
- DP2OD004437/OD/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources