Characterization of microRNAs involved in embryonic stem cell states - PubMed (original) (raw)
doi: 10.1089/scd.2009.0426.
Irena Ivanovska, Kshama Mehta, Sunny Song, Angelique Nelson, Yunbing Tan, Julie Mathieu, Christopher Darby, C Anthony Blau, Carol Ware, Garrick Peters, Daniel G Miller, Lanlan Shen, Michele A Cleary, Hannele Ruohola-Baker
Affiliations
- PMID: 20128659
- PMCID: PMC3128320
- DOI: 10.1089/scd.2009.0426
Characterization of microRNAs involved in embryonic stem cell states
Bradford Stadler et al. Stem Cells Dev. 2010 Jul.
Abstract
Studies of embryonic stem cells (ESCs) reveal that these cell lines can be derived from differing stages of embryonic development. We analyzed common changes in the expression of microRNAs (miRNAs) and mRNAs in 9 different human ESC (hESC) lines during early commitment and further examined the expression of key ESCenriched miRNAs in earlier developmental states in several species. We show that several previously defined hESC-enriched miRNA groups (the miR-302, -17, and -515 families, and the miR-371-373 cluster) and several other hESC-enriched miRNAs are down-regulated rapidly in response to differentiation. We further found that mRNAs up-regulated upon differentiation are enriched in potential target sites for these hESC-enriched miRNAs. Interestingly, we also observed that the expression of ESC-enriched miRNAs bearing identical seed sequences changed dynamically while the cells transitioned through early embryonic states. In human and monkey ESCs, as well as human-induced pluripotent stem cells (iPSCs), the miR-371-373 cluster was consistently up-regulated, while the miR-302 family was mildly down-regulated when the cells were chemically treated to regress to an earlier developmental state. Similarly, miR-302b, but not mmu-miR-295, was expressed at higher levels in murine epiblast stem cells (mEpiSC) as compared with an earlier developmental state, mouse ESCs. These results raise the possibility that the relative expression of related miRNAs might serve as diagnostic indicators in defining the developmental state of embryonic cells and other stem cell lines, such as iPSCs. These data also raise the possibility that miRNAs bearing identical seed sequences could have specific functions during separable stages of early embryonic development.
Figures
FIG. 1.
Assessing undirected differentiation in human embryonic stem cells (hESCs). (A) Undifferentiated hESCs were treated with either mouse embryonic fibroblast (MEF)-conditioned media to maintain an undifferentiated state or serum-containing media to induce undirected differentiation over a 4-day period. Total RNA was extracted from cells and used for qRT-PCR, microRNA (miRNA), and mRNA microarray analysis. (B) Stem cell marker expression levels at Day 4 of the differentiation scheme were assessed by both qRT-PCR and microarray in all undifferentiated and differentiated hESC lines. qRT-PCR analysis was performed as described in B, and both microarray and qRT-PCR data are expressed as the change in the level of gene expression during 4-day differentiation. (C) Phase-contrast microscopy was used to assess morphology of hESC colonies at Day 3 of the differentiation scheme. All images shown were captured using 10× magnification. (D) Immunofluorescent detection of the stem cell markers Oct4, SSEA-4, and Tra-1–60 demonstrates a strong reduction in stem cell markers in the differentiating H1 hESCs. Images were captured using confocal microscopy at 40× magnification.
FIG. 2.
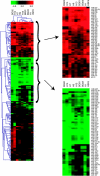
Clustering analysis of microRNA (miRNA) expression reveals human embryonic stem cell (hESC)-enriched miRNAs. Single-color, miRNA microarrays (Agilent Technologies, Santa Clara, CA) were used to analyze miRNA expression in 9 hESC lines. Of the 470 miRNAs detected by the array, 184 were consistently expressed above background levels in the samples and were included in the clustering analysis. The log2 ratio of differentiated/undifferentiated signal (fold change) for each cell line was subjected to hierarchical clustering by Euclidean distance metric using average linkage. miRNAs that are down-regulated upon differentiation are indicated by green color in the cluster and up-regulated miRNAs are indicated by red.
FIG. 3.
Genome-wide mRNA profiling and differential gene expression analysis in human embryonic stem cell (hESC) lines. (A) Two-way hierarchical clustering of whole genome microarray expression (Affymetryx) was used to find groups of genes whose expression changed at least 1.5-fold and had a P value <0.01 in all 9 hESCs. This analysis generated a group of 1,263 genes down-regulated (blue) and 1,509 genes up-regulated (pink). (B) The gene set of up-regulated mRNAs generated from the clustering analysis was analyzed for enrichment of biological function using gene ontology (GO) biological process functional categories and analyzed for enrichment of hexamer sequences in their 3′-UTRs. (C) The gene set of down-regulated mRNAs generated from the clustering analysis was annotated by GO biological process functional categories and analyzed for enrichment of hexamer sequences in their 3′-UTRs as described in B.
FIG. 4.
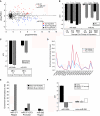
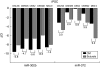
Relative expression of microRNAs (miRNAs) with similar seeds delineates separable embryonic states. (A) Microarray data for a BG02 human embryonic stem cell (hESC) line cultured in conditioned media (CM) or low-level sodium butyrate media (butyrate) as depicted by scatter plot. A Bayesian approach was utilized to determine significant genes as described in Materials and Methods, and the data are plotted as the log ratio (Buty/CM) versus the log average intensity (Buty+CM) for each miRNA gene. (B) qPCR data for the miR-302b and miR-372 are shown for hESC lines cultured in CM or butyrate media. Two independent pairs of samples are shown for both the BG02 and H13 hESC lines, as indicated by the dashed error bars. Data are represented as ΔCt (average Ct of miRNA – average Ct of RNU66 control RNA), and fold changes (butyrate/CM) in expression are indicated below each pair of samples. (C) Nonhuman primate MF-1 cells were maintained on feeder cells in ES media or exposed to sodium butyrate-containing media and qPCR analysis for miR-302b and miR-372 was performed. Data are represented as ΔCt (miRNA-RNU66 control RNA) and fold changes (butyrate/ES media) in expression are indicated below the samples. (D) ChIP assays were performed in conjunction with DNA microarray analysis to determine promoter regions enriched for acetylated histone, H3K9, a mark of active chromatin. The region of chromosome 19 containing the hsa-miR-371–373 locus is displayed and a fold enrichment of the ac-H9K3 antibody in comparison with the input fractions for both CM-treated and butyrate-treated cells are shown. (E) ChIP using an antibody specific for acetylated H3K9 was performed and subsequent qPCR for specific miR-371–373 promoter and mature miRNA regions (both miR-372 and miR-302) were performed from both CM and butyrate-treated extracts. Data are normalized to control samples obtained from ChIP with a histone H3 antibody. (F) qPCR analysis of murine mmu-miR-295 and miR-302b are shown for mESC and mEpiSC. Both naturally derived mEpiSC and chemically induced EpiSC cell lines were analyzed. The data are represented as ΔCt (miRNA-snoRNA202).
FIG. 5.
Similar to human embryonic stem cells (hESCs) regressed to earlier stem cell states, induced pluripotent stem cells (iPSCs) show increased miR-372 levels upon butyrate treatment. Five distinct iPSC clonal cell lines were generated and validated as described in Materials and Methods. qPCR data for the miR-302b and miR-372 are shown for these iPSC lines cultured in conditioned media (CM) or butyrate-containing media (butyrate). Data are represented as ΔCt (miRNA-RNU66 control RNA), and fold changes (butyrate/CM) in expression are indicated below each pair of samples.
FIG. 6.
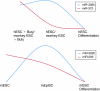
MicroRNAs (miRNAs) with identical seed sequences are differentially expressed at separable stages of embryonic development. Summary of the observed changes in miR-302b and hsa-miR-372/mmu-miR-295 expression in early embryonic stages and in response to differentiation. The highest levels of miR-372/mmu-miR-295 are found at the earliest stages in butyrate-treated primate embryonic stem cells (ES cells) and in murine ES cells, while miR-302b levels increase at a later embryonic stage represented by primate ES cells and murine epiSCs.
Similar articles
- Distinct MicroRNA Expression Signatures of Porcine Induced Pluripotent Stem Cells under Mouse and Human ESC Culture Conditions.
Zhang W, Zhong L, Wang J, Han J. Zhang W, et al. PLoS One. 2016 Jul 6;11(7):e0158655. doi: 10.1371/journal.pone.0158655. eCollection 2016. PLoS One. 2016. PMID: 27384321 Free PMC article. - Identification of microRNAs regulated by activin A in human embryonic stem cells.
Tsai ZY, Singh S, Yu SL, Kao LP, Chen BZ, Ho BC, Yang PC, Li SS. Tsai ZY, et al. J Cell Biochem. 2010 Jan 1;109(1):93-102. doi: 10.1002/jcb.22385. J Cell Biochem. 2010. PMID: 19885849 - Embryonic stem cell microRNAs: defining factors in induced pluripotent (iPS) and cancer (CSC) stem cells?
Gunaratne PH. Gunaratne PH. Curr Stem Cell Res Ther. 2009 Sep;4(3):168-77. doi: 10.2174/157488809789057400. Curr Stem Cell Res Ther. 2009. PMID: 19492978 Review. - Vitamin C induces a pluripotent state in mouse embryonic stem cells by modulating microRNA expression.
Gao Y, Han Z, Li Q, Wu Y, Shi X, Ai Z, Du J, Li W, Guo Z, Zhang Y. Gao Y, et al. FEBS J. 2015 Feb;282(4):685-99. doi: 10.1111/febs.13173. Epub 2015 Jan 8. FEBS J. 2015. PMID: 25491368 - The Key Role of MicroRNAs in Self-Renewal and Differentiation of Embryonic Stem Cells.
Divisato G, Passaro F, Russo T, Parisi S. Divisato G, et al. Int J Mol Sci. 2020 Aug 31;21(17):6285. doi: 10.3390/ijms21176285. Int J Mol Sci. 2020. PMID: 32877989 Free PMC article. Review.
Cited by
- Analysis of CDS-located miRNA target sites suggests that they can effectively inhibit translation.
Hausser J, Syed AP, Bilen B, Zavolan M. Hausser J, et al. Genome Res. 2013 Apr;23(4):604-15. doi: 10.1101/gr.139758.112. Epub 2013 Jan 18. Genome Res. 2013. PMID: 23335364 Free PMC article. - Developmentally programmed 3' CpG island methylation confers tissue- and cell-type-specific transcriptional activation.
Yu DH, Ware C, Waterland RA, Zhang J, Chen MH, Gadkari M, Kunde-Ramamoorthy G, Nosavanh LM, Shen L. Yu DH, et al. Mol Cell Biol. 2013 May;33(9):1845-58. doi: 10.1128/MCB.01124-12. Epub 2013 Mar 4. Mol Cell Biol. 2013. PMID: 23459939 Free PMC article. - The Wnt/β-catenin pathway regulates the expression of the miR-302 cluster in mouse ESCs and P19 cells.
Bräutigam C, Raggioli A, Winter J. Bräutigam C, et al. PLoS One. 2013 Sep 10;8(9):e75315. doi: 10.1371/journal.pone.0075315. eCollection 2013. PLoS One. 2013. PMID: 24040406 Free PMC article. - Downregulation of ULK1 by microRNA-372 inhibits the survival of human pancreatic adenocarcinoma cells.
Chen H, Zhang Z, Lu Y, Song K, Liu X, Xia F, Sun W. Chen H, et al. Cancer Sci. 2017 Sep;108(9):1811-1819. doi: 10.1111/cas.13315. Epub 2017 Aug 10. Cancer Sci. 2017. PMID: 28677209 Free PMC article. - mRNA and miRNA expression profiles in an ectoderm-biased substate of human pluripotent stem cells.
Mawaribuchi S, Aiki Y, Ikeda N, Ito Y. Mawaribuchi S, et al. Sci Rep. 2019 Aug 15;9(1):11910. doi: 10.1038/s41598-019-48447-z. Sci Rep. 2019. PMID: 31417139 Free PMC article.
References
- Thomson JA. Itskovitz-Eldor J. Shapiro SS. Waknitz MA. Swiergiel JJ. Marshall VS. Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. - PubMed
- Evans MJ. Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. - PubMed
- Klimanskaya I. Rosenthal N. Lanza R. Derive and conquer: sourcing and differentiating stem cells for therapeutic applications. Nat Rev Drug Discov. 2008;7:131–142. - PubMed
- Spagnoli FM. Hemmati-Brivanlou A. Guiding embryonic stem cells towards differentiation: lessons from molecular embryology. Curr Opin Genet Dev. 2006;16:469–475. - PubMed
- Maherali N. Sridharan R. Xie W. Utikal J. Eminli S. Arnold K. Stadtfeld M. Yachechko R. Tchieu J. Jaenisch R. Plath K. Hochedlinger K. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases