The CRISPR system: small RNA-guided defense in bacteria and archaea - PubMed (original) (raw)
Review
The CRISPR system: small RNA-guided defense in bacteria and archaea
Fedor V Karginov et al. Mol Cell. 2010.
Abstract
All cellular systems evolve ways to combat predators and genomic parasites. In bacteria and archaea, numerous resistance mechanisms have developed against phage. Our understanding of this defensive repertoire has recently been expanded to include the CRISPR system of clustered, regularly interspaced short palindromic repeats. In this remarkable pathway, short sequence tags from invading genetic elements are actively incorporated into the host's CRISPR locus to be transcribed and processed into a set of small RNAs that guide the destruction of foreign genetic material. Here we review the inner workings of this adaptable and heritable immune system and draw comparisons to small RNA-guided defense mechanisms in eukaryotic cells.
Copyright 2010 Elsevier Inc. All rights reserved.
Figures
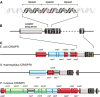
Figure 1. The structure of a CRISPR locus
(A) Repeat sequences averaging 32bp are interleaved by variable spacers of approximately the same size. (B) The number of repeat-spacer units varies greatly. A conserved leader sequence (gray) of several hundred base pairs is located on one side of the cluster. (C) CRISPR-associated (cas) genes surround the CRISPR locus. Three examples of well-studied CRISPR loci are shown. Core cas genes are depicted in red, subtype-specific genes in blue, and the RAMP module in green. Unclassified genes are shown in dark grey. Gene names follow the nomenclature of Haft et al., except cas7, which was named by Barrangou et al.
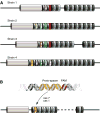
Figure 2. Diversity and acquisition of spacer content
(A) A schematic representation of the CRISPR loci in related bacterial strains. Colored spacers represent sequences that differ among strains. All strains share the same ancestral spacers at the leader-distal end (gray spacers). Deletions after divergence have occurred in some strains. Spacer content becomes more strain-specific near the leader end. Strain ancestry can be traced by the presence of common spacers (green, red). (B) Acquisition of new spacers occurs next to the leader sequence. The spacer sequence is derived from the invading nucleic acid, selected based upon the presence of a proto-spacer adjacent motif.
Figure 3. Processing of CRISPR content into crRNAs
The locus is transcribed from the leader sequence and the RNA is cleaved within the repeat by cse3 or cas6. An additional processing step yields the mature crRNA/psiRNA consisting of an 8nt repeat tag and the spacer sequence.
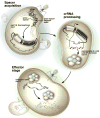
Figure 4. An overall model of CRISPR/cas activity
During spacer acquisition, sequence elements from invading nucleic acids become incorporated at the leader-proximal end of the CRISPR locus. In the processing stage, the locus is transcribed and processed into mature crRNA/psiRNAs containing an 8nt repeat tag and a single spacer unit. During the effector stage, the mature crRNAs in complex with associated cas proteins leads to degradation of complementary nucleic acids. See text for details.
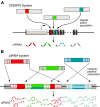
Figure 5. Conserved themes in small RNA-guided defense
(A) The CRISPR system actively incorporates short sequence tags from invading nucleic acids into the CRISPR locus of regularly interspaced repeats. (B) In the piRNA system, genomic piRNA clusters passively acquire new content by movement of transposons into the locus. In both cases, the locus is transcribed and processed into small RNAs that guide the destruction of exogenous or parasitic sequences.
Similar articles
- In defense of phage: viral suppressors of CRISPR-mediated adaptive immunity in bacteria.
Wiedenheft B. Wiedenheft B. RNA Biol. 2013 May;10(5):886-90. doi: 10.4161/rna.23591. RNA Biol. 2013. PMID: 23392292 Free PMC article. - CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea.
Marraffini LA, Sontheimer EJ. Marraffini LA, et al. Nat Rev Genet. 2010 Mar;11(3):181-90. doi: 10.1038/nrg2749. Nat Rev Genet. 2010. PMID: 20125085 Free PMC article. Review. - CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation.
Bhaya D, Davison M, Barrangou R. Bhaya D, et al. Annu Rev Genet. 2011;45:273-97. doi: 10.1146/annurev-genet-110410-132430. Annu Rev Genet. 2011. PMID: 22060043 Review. - CRISPR/Cas, the immune system of bacteria and archaea.
Horvath P, Barrangou R. Horvath P, et al. Science. 2010 Jan 8;327(5962):167-70. doi: 10.1126/science.1179555. Science. 2010. PMID: 20056882 Review. - CRISPR-mediated adaptive immune systems in bacteria and archaea.
Sorek R, Lawrence CM, Wiedenheft B. Sorek R, et al. Annu Rev Biochem. 2013;82:237-66. doi: 10.1146/annurev-biochem-072911-172315. Epub 2013 Mar 11. Annu Rev Biochem. 2013. PMID: 23495939 Review.
Cited by
- Draft genome sequence of Ureibacillus thermosphaericus strain thermo-BF, isolated from Ramsar hot springs in Iran.
Abbasalizadeh S, Salehi Jouzani G, Motamedi Juibari M, Azarbaijani R, Parsa Yeganeh L, Ahmad Raji M, Mardi M, Salekdeh GH. Abbasalizadeh S, et al. J Bacteriol. 2012 Aug;194(16):4431. doi: 10.1128/JB.00634-12. J Bacteriol. 2012. PMID: 22843574 Free PMC article. - devI is an evolutionarily young negative regulator of Myxococcus xanthus development.
Rajagopalan R, Wielgoss S, Lippert G, Velicer GJ, Kroos L. Rajagopalan R, et al. J Bacteriol. 2015 Apr;197(7):1249-62. doi: 10.1128/JB.02542-14. Epub 2015 Feb 2. J Bacteriol. 2015. PMID: 25645563 Free PMC article. - Molecular Marvels: Small Molecules Paving the Way for Enhanced Gene Therapy.
Hasselbeck S, Cheng X. Hasselbeck S, et al. Pharmaceuticals (Basel). 2023 Dec 27;17(1):41. doi: 10.3390/ph17010041. Pharmaceuticals (Basel). 2023. PMID: 38256875 Free PMC article. Review. - Comparison of Intracellular "Ca. Endomicrobium Trichonymphae" Genomovars Illuminates the Requirement and Decay of Defense Systems against Foreign DNA.
Izawa K, Kuwahara H, Kihara K, Yuki M, Lo N, Itoh T, Ohkuma M, Hongoh Y. Izawa K, et al. Genome Biol Evol. 2016 Oct 13;8(10):3099-3107. doi: 10.1093/gbe/evw227. Genome Biol Evol. 2016. PMID: 27635050 Free PMC article. - Adaptation and modification of three CRISPR loci in two closely related cyanobacteria.
Hein S, Scholz I, Voß B, Hess WR. Hein S, et al. RNA Biol. 2013 May;10(5):852-64. doi: 10.4161/rna.24160. Epub 2013 Mar 27. RNA Biol. 2013. PMID: 23535141 Free PMC article.
References
- Agari Y, Sakamoto K, Tamakoshi M, Oshima T, Kuramitsu S, Shinkai A. Transcription Profile of Thermus thermophilus CRISPR Systems after Phage Infection. J Mol Biol 2009 - PubMed
- Agari Y, Yokoyama S, Kuramitsu S, Shinkai A. X-ray crystal structure of a CRISPR-associated protein, Cse2, from Thermus thermophilus HB8. Proteins. 2008;73:1063–1067. - PubMed
- Andersson AF, Banfield JF. Virus population dynamics and acquired virus resistance in natural microbial communities. Science. 2008;320:1047–1050. - PubMed
- Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. - PubMed
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases