Recognition of histone H3K4 trimethylation by the plant homeodomain of PHF2 modulates histone demethylation - PubMed (original) (raw)
Recognition of histone H3K4 trimethylation by the plant homeodomain of PHF2 modulates histone demethylation
Hong Wen et al. J Biol Chem. 2010.
Abstract
Distinct lysine methylation marks on histones create dynamic signatures deciphered by the "effector" modules, although the underlying mechanisms remain unclear. We identified the plant homeodomain- and Jumonji C domain-containing protein PHF2 as a novel histone H3K9 demethylase. We show in biochemical and crystallographic analyses that PHF2 recognizes histone H3K4 trimethylation through its plant homeodomain finger and that this interaction is essential for PHF2 occupancy and H3K9 demethylation at rDNA promoters. Our study provides molecular insights into the mechanism by which distinct effector domains within a protein cooperatively modulate the "cross-talk" of histone modifications.
Figures
FIGURE 1.
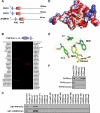
The PHF2 PHD finger specifically binds to histone H3K4me2/3. A, schematic representation of human PHF2 family proteins. B, the PHF2 PHD finger binds to H3K4me2/3 in a histone peptide array. Note that H3 (amino acids (aa) 44–64) peptides showed nonspecific binding. C, Western analysis of PHF2 and JHDM1D PHD fingers in peptide pulldown assays. D, crystal structure of the PHF2 PHD finger in complex with an H3K4me3 peptide. The PHD finger is shown as electrostatic surface, with red as negatively charged surface and blue as positively charged surface, and the histone peptide is shown as a stick, with carbon, oxygen, and nitrogen atoms colored yellow, red, and blue, respectively. E, stick representation of H3K4me3 (yellow) in the aromatic cage of the PHF2 PHD finger composed of Tyr7, Tyr14, Met20, and Trp29 residues (green). D–E were generated using PyMOL. F, Western analysis of peptide pulldown assays as described in C with PHF2 PHD finger mutants Y7A and W29A.
FIGURE 2.
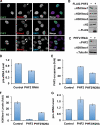
The PHD finger is important for PHF2-mediated promoter demethylation and expression of rDNA. A and B, PHF2 specifically demethylates H3K9me1 in vivo. A, immunostaining of U2OS cells expressing green fluorescent protein-tagged PHF2 with the indicated anti-H3K9me antibodies and 4′,6-diamidino-2-phenylindole (DAPI). Cells expressing ectopic PHF2 are indicated with arrows. B, Western analysis with the indicated antibodies of whole cell extracts from 293T cells transfected with FLAG-PHF2. Ectopic PHF2 was detected with anti-FLAG antibody. Histone H3 is shown as a loading control. C, knockdown of PHF2 increases the H3K9me1 level. Shown are the results from Western analysis performed with endogenous PHF2 and total H3K9me1 levels in whole cell extract from 293T cells expressing control or PHF2 shRNA. Tubulin is shown as a loading control. RNAi, RNA interference. D, knockdown of PHF2 decreases pre-rRNA levels. Shown are the results from real-time PCR analysis of pre-rRNA levels in 293T cells expressing control or PHF2 shRNA. E, the PHD finger is required for PHF2 occupancy at the rDNA promoter. Shown are the results from real-time PCR analysis of the rDNA promoter for PHF2 ChIP samples from cells expressing PHF2 or PHF2(W29A). ChIP signals are shown as -fold relative to IgG ChIPs. F, the PHD finger is required for PHF2-mediated H3K9 demethylation at the rDNA promoter. Shown are the results from real-time PCR analysis of the rDNA promoter for H3K9me1 ChIP samples as described for E. G, mutation in the PHD finger attenuates PHF2-dependent activation of rDNA expression. Shown are the results from real-time PCR analysis of pre-rRNA levels in 293T cells as in described for E. In D–G, error bars represent S.E., and all p values are <0.05.
Similar articles
- PHD finger protein 2 (PHF2) represses ribosomal RNA gene transcription by antagonizing PHF finger protein 8 (PHF8) and recruiting methyltransferase SUV39H1.
Shi G, Wu M, Fang L, Yu F, Cheng S, Li J, Du JX, Wong J. Shi G, et al. J Biol Chem. 2014 Oct 24;289(43):29691-700. doi: 10.1074/jbc.M114.571653. Epub 2014 Sep 9. J Biol Chem. 2014. PMID: 25204660 Free PMC article. - The histone demethylase PHF2 promotes fat cell differentiation as an epigenetic activator of both C/EBPα and C/EBPδ.
Lee KH, Ju UI, Song JY, Chun YS. Lee KH, et al. Mol Cells. 2014 Oct 31;37(10):734-41. doi: 10.14348/molcells.2014.0180. Epub 2014 Sep 29. Mol Cells. 2014. PMID: 25266703 Free PMC article. - A complete methyl-lysine binding aromatic cage constructed by two domains of PHF2.
Horton JR, Zhou J, Chen Q, Zhang X, Bedford MT, Cheng X. Horton JR, et al. J Biol Chem. 2023 Feb;299(2):102862. doi: 10.1016/j.jbc.2022.102862. Epub 2022 Dec 31. J Biol Chem. 2023. PMID: 36596360 Free PMC article. - Plant homeodomain fingers form a helping hand for transcription.
Fortschegger K, Shiekhattar R. Fortschegger K, et al. Epigenetics. 2011 Jan;6(1):4-8. doi: 10.4161/epi.6.1.13297. Epub 2011 Jan 1. Epigenetics. 2011. PMID: 20818169 Free PMC article. Review. - A Structural Perspective on Readout of Epigenetic Histone and DNA Methylation Marks.
Patel DJ. Patel DJ. Cold Spring Harb Perspect Biol. 2016 Mar 1;8(3):a018754. doi: 10.1101/cshperspect.a018754. Cold Spring Harb Perspect Biol. 2016. PMID: 26931326 Free PMC article. Review.
Cited by
- Decreased SUV39H1 at the promoter region leads to increased CREMα and accelerates autoimmune response in CD4+ T cells from patients with systemic lupus erythematosus.
Luo S, Zhang H, Xie Y, Huang J, Luo D, Zhang Q. Luo S, et al. Clin Epigenetics. 2022 Dec 20;14(1):181. doi: 10.1186/s13148-022-01411-7. Clin Epigenetics. 2022. PMID: 36536372 Free PMC article. - The PHD finger: a versatile epigenome reader.
Sanchez R, Zhou MM. Sanchez R, et al. Trends Biochem Sci. 2011 Jul;36(7):364-72. doi: 10.1016/j.tibs.2011.03.005. Epub 2011 Apr 21. Trends Biochem Sci. 2011. PMID: 21514168 Free PMC article. Review. - Plant homeodomain (PHD) fingers of CHD4 are histone H3-binding modules with preference for unmodified H3K4 and methylated H3K9.
Mansfield RE, Musselman CA, Kwan AH, Oliver SS, Garske AL, Davrazou F, Denu JM, Kutateladze TG, Mackay JP. Mansfield RE, et al. J Biol Chem. 2011 Apr 1;286(13):11779-91. doi: 10.1074/jbc.M110.208207. Epub 2011 Jan 28. J Biol Chem. 2011. PMID: 21278251 Free PMC article. - Inhibitors of Protein Methyltransferases and Demethylases.
Kaniskan HÜ, Martini ML, Jin J. Kaniskan HÜ, et al. Chem Rev. 2018 Feb 14;118(3):989-1068. doi: 10.1021/acs.chemrev.6b00801. Epub 2017 Mar 24. Chem Rev. 2018. PMID: 28338320 Free PMC article. Review. - Targeting Histone Modifications in Breast Cancer: A Precise Weapon on the Way.
Li W, Wu H, Sui S, Wang Q, Xu S, Pang D. Li W, et al. Front Cell Dev Biol. 2021 Sep 14;9:736935. doi: 10.3389/fcell.2021.736935. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34595180 Free PMC article. Review.
References
- Berger S. L. (2007) Nature 447, 407–412 - PubMed
- Kouzarides T. (2007) Cell 128, 693–705 - PubMed
- Margueron R., Trojer P., Reinberg D. (2005) Curr. Opin. Genet. Dev. 15, 163–176 - PubMed
- Shilatifard A. (2006) Annu. Rev. Biochem. 75, 243–269 - PubMed
- Li B., Carey M., Workman J. L. (2007) Cell 128, 707–719 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases