Gene ontology analysis for RNA-seq: accounting for selection bias - PubMed (original) (raw)
Gene ontology analysis for RNA-seq: accounting for selection bias
Matthew D Young et al. Genome Biol. 2010.
Abstract
We present GOseq, an application for performing Gene Ontology (GO) analysis on RNA-seq data. GO analysis is widely used to reduce complexity and highlight biological processes in genome-wide expression studies, but standard methods give biased results on RNA-seq data due to over-detection of differential expression for long and highly expressed transcripts. Application of GOseq to a prostate cancer data set shows that GOseq dramatically changes the results, highlighting categories more consistent with the known biology.
Figures
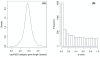
Figure 1
Length distribution of genes in Gene Ontology categories. (a) The distribution of average gene lengths in GO categories on a log10 scale. The GO category gene length is given by the median length of the genes within the category. (b) _P_-values for the two-sided Mann-Whitney U test comparing the median length of genes in a GO category with the overall distribution of genes for 7,873 GO categories. The excess of low _P_-values shows that there are many GO categories that contain a set of significantly long or short genes.
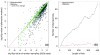
Figure 2
Differential expression as a function of gene length and read count. (a) The proportion of DE genes is plotted as a function of the transcript length. Each point represents the percentage of genes called DE in a bin of 300 genes plotted against the median gene length for the bin. The green line is the probability weighting function obtained by fitting a monotonic cubic spline with 6 knots to the binary data. A clear trend towards more detected differential expression at longer gene lengths can be seen. (b) The same, except instead of transcript length, the total number of reads for each gene was used. Again, a trend towards more DE for genes with more reads can be seen. Note the greater range of probabilities compared to (a).
Figure 3
Change in Gene Ontology category rank between the standard and GOseq methodologies. (a) Change in rank of GO categories going from the hypergeometric method to GOseq correcting for length bias plotted against the log of the average gene length of the category. (b) Change in rank of GO categories going from the hypergeometric method to GOseq correcting for total read count plotted against the log of the average number of counts of each gene in the category. A trend for the standard method to underestimate significance for GO categories containing short (or highly expressed) genes and overestimate significance for GO categories containing long (or underexpressed) genes can be clearly seen.
Figure 4
Comparison of GOseq and the standard hypergeometric methods. (a) The _P_-values generated with GOseq using the Wallenius approximation and the standard hypergeometric method are plotted against the _P_-values calculated with GOseq using random sampling (200k repeats). The Wallenius method (green crosses) shows good agreement with the high resolution (200,000 repeats) random sampling. A large discrepancy in _P_-values is seen between GOseq and the hypergeometric method. (a) The number of discrepancies between lists is shown for a given list size. The black line compares GOseq using high resolution sampling with the hypergeometric method. The red line compares GOseq using high resolution sampling with the Wallenius approximation. Again, GOseq using the Wallenius method shows little difference from GOseq using the random sampling method (with 200k repeats) while the hypergeometric method shows a large number (approximately 20%) of discrepancies.
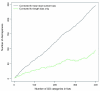
Figure 5
A comparison of Gene Ontology analysis using RNA-seq and microarrays on the same samples. The fraction of GO categories identified by RNA-seq data that overlap with the microarray GO analysis are shown as a function of the number of categories selected. RNA-seq data have been analyzed using GOseq and hypergeometric methods. The GOseq categories have a consistently higher overlap with the microarray GO categories than the standard method.
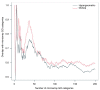
Figure 6
Change in most significant categories when correcting for total read count bias or length bias versus the standard method. A plot of the number of discrepancies in the most significant GO categories generated using different methods. This plot compares the length bias correcting version of GOseq to the standard hypergeometric method (green line) and the total read count bias correcting version of GO-seq to the standard hypergeometric method (black line).
Similar articles
- GSEPD: a Bioconductor package for RNA-seq gene set enrichment and projection display.
Stamm K, Tomita-Mitchell A, Bozdag S. Stamm K, et al. BMC Bioinformatics. 2019 Mar 6;20(1):115. doi: 10.1186/s12859-019-2697-5. BMC Bioinformatics. 2019. PMID: 30841846 Free PMC article. - A Workflow Guide to RNA-seq Analysis of Chaperone Function and Beyond.
Lang BJ, Holton KM, Gong J, Calderwood SK. Lang BJ, et al. Methods Mol Biol. 2018;1709:233-252. doi: 10.1007/978-1-4939-7477-1_18. Methods Mol Biol. 2018. PMID: 29177664 Free PMC article. - Analysis of androgen and anti-androgen regulation of KLK-related peptidase 2, 3, and 4 alternative transcripts in prostate cancer.
Lai J, An J, Nelson CC, Lehman ML, Batra J, Clements JA. Lai J, et al. Biol Chem. 2014 Sep;395(9):1127-32. doi: 10.1515/hsz-2014-0149. Biol Chem. 2014. PMID: 25153393 - Gene dispersion is the key determinant of the read count bias in differential expression analysis of RNA-seq data.
Yoon S, Nam D. Yoon S, et al. BMC Genomics. 2017 May 25;18(1):408. doi: 10.1186/s12864-017-3809-0. BMC Genomics. 2017. PMID: 28545404 Free PMC article. - Uncovering the complexity of transcriptomes with RNA-Seq.
Costa V, Angelini C, De Feis I, Ciccodicola A. Costa V, et al. J Biomed Biotechnol. 2010;2010:853916. doi: 10.1155/2010/853916. Epub 2010 Jun 27. J Biomed Biotechnol. 2010. PMID: 20625424 Free PMC article. Review.
Cited by
- Gene Ontology Meta Annotator for Plants (GOMAP).
Wimalanathan K, Lawrence-Dill CJ. Wimalanathan K, et al. Plant Methods. 2021 May 25;17(1):54. doi: 10.1186/s13007-021-00754-1. Plant Methods. 2021. PMID: 34034755 Free PMC article. - Effect of Fat Content on Rice Taste Quality through Transcriptome Analysis.
Guo J, Zhou X, Chen D, Chen K, Ye C, Liu J, Liu S, Chen Y, Chen G, Liu C. Guo J, et al. Genes (Basel). 2024 Jan 9;15(1):81. doi: 10.3390/genes15010081. Genes (Basel). 2024. PMID: 38254970 Free PMC article. - Temporal and Spatial Signatures of Scylla paramamosain Transcriptome Reveal Mechanistic Insights into Endogenous Ovarian Maturation under Risk of Starvation.
Fu Y, Zhang F, Wang W, Xu J, Zhao M, Ma C, Cheng Y, Chen W, Su Z, Lv X, Liu Z, Ma K, Ma L. Fu Y, et al. Int J Mol Sci. 2024 Jan 5;25(2):700. doi: 10.3390/ijms25020700. Int J Mol Sci. 2024. PMID: 38255774 Free PMC article. - Argonaute-CLIP delineates versatile, functional RNAi networks in Aedes aegypti, a major vector of human viruses.
Rozen-Gagnon K, Gu M, Luna JM, Luo JD, Yi S, Novack S, Jacobson E, Wang W, Paul MR, Scheel TKH, Carroll T, Rice CM. Rozen-Gagnon K, et al. Cell Host Microbe. 2021 May 12;29(5):834-848.e13. doi: 10.1016/j.chom.2021.03.004. Epub 2021 Mar 31. Cell Host Microbe. 2021. PMID: 33794184 Free PMC article. - Human DDX6 regulates translation and decay of inefficiently translated mRNAs.
Weber R, Chang CT. Weber R, et al. Elife. 2024 Jul 11;13:RP92426. doi: 10.7554/eLife.92426. Elife. 2024. PMID: 38989862 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases