Heterogeneity in spike protein genes of porcine epidemic diarrhea viruses isolated in Korea - PubMed (original) (raw)
Heterogeneity in spike protein genes of porcine epidemic diarrhea viruses isolated in Korea
Dong-Kyu Lee et al. Virus Res. 2010 May.
Abstract
Porcine epidemic diarrhea virus (PEDV) has plagued the domestic swine industry in Korea causing significant economic impacts on pig production nationwide. In the present study, we determined the complete nucleotide sequences of the spike (S) glycoprotein genes of seven Korean PEDV isolates. The entire S genes of all isolates were found to be nine nucleotides longer in length than other PEDV reference strains. This size difference was due to the combined presence of notable 15 bp insertion and 6 bp deletion within the N-terminal region of the S1 domain of the Korean isolates. In addition, the largest number of amino acid variations was accumulated in the S1 N-terminal region, leading to the presence of hypervariability in the isolates. Sequence comparisons at the peptide level of the S proteins revealed that all seven Korean isolates shared diverse similarities ranging from a 93.6% to 99.6% identity with each other but exhibited a 92.2% to 93.7% identity with other reference strains. Collectively, the sequence analysis data indicate the diversity of the PEDV isolates currently prevalent in Korea that represents a heterogeneous group. Phylogenetic analyses showed two separate clusters, in which all Korean field isolates were grouped together in the second cluster (group 2). The results indicate that prevailing isolates in Korea are phylogenetically more closely related to each other rather than other reference strains. Interestingly, the tree topology based on the nucleotide sequences representing the S1 domain or the S1 N-terminal region most nearly resembled the full S gene-based phylogenetic tree. Therefore, our data implicates a potential usefulness of the partial S protein gene including the N-terminal region in unveiling genetic relatedness of PEDV isolates.
(c) 2010 Elsevier B.V. All rights reserved.
Figures
Fig. 1
Amino acid sequence alignment of the S glycoprotein genes of the Korean PEDV isolates and PEDV reference strains. The dashes (–) indicate the deleted sequences. Potential _N_-glycosylation sites predicted by GlycoMod Tool (
http://www.expasy.ch/tools/glycomod/
) are shown in boldface type. Insertions and deletions in PEDV isolates are shaded. Putative signal sequences (dotted line box) and amino acids representing potential hypervariable domains (solid boxes) are indicated.
Fig. 1
Amino acid sequence alignment of the S glycoprotein genes of the Korean PEDV isolates and PEDV reference strains. The dashes (–) indicate the deleted sequences. Potential _N_-glycosylation sites predicted by GlycoMod Tool (
http://www.expasy.ch/tools/glycomod/
) are shown in boldface type. Insertions and deletions in PEDV isolates are shaded. Putative signal sequences (dotted line box) and amino acids representing potential hypervariable domains (solid boxes) are indicated.
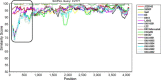
Fig. 2
Similarity plot of the full-length nucleotide sequences of seven PEDV isolates with other reference strains. The similarity plot was constructed using the two-parameter (Kimura) distance model with a sliding window of 200 bp and step size of 20 bp. The vertical and horizontal axes indicate the percent nucleotide similarity and nucleotide position (bp) in the graph, respectively.
Fig. 3
Phylogenetic analyses based on the nucleotide sequences corresponding to the S1 C-terminal region (495–697 aa) (A), full-length S gene (B), S1 domain (C), S1 N-terminal region (1–494 aa) (D), and S2 domain (736–1383 aa) (E). Putative similar regions of spike proteins of other distantly related coronaviruses, transmissible gastroenteritis virus (TGEV) and human coronavirus HCoV-229E, were also included in this study. Multiple-sequencing alignments were performed using ClustalX program and the phylogenetic tree was constructed from the aligned nucleotide sequences by using the neighbor-joining method. The numbers at each branch represent bootstrap values greater than 50% of 1000 replicates. The scale bars indicate the number of 0.1 inferred substitutions per site.
References
- Chang S.H., Bae J.L., Kang T.J., Kim J., Chung G.H., Lim C.W., Laude H., Yang M.S., Jang Y.S. Identification of the epitope region capable of inducing neutralizing antibodies against the porcine epidemic diarrhea virus. Mol. Cells. 2002;14:295–299. - PubMed
- Debouck P., Pensaert M. Experimental infection of pigs with a new porcine enteric coronavirus, CV777. Am. J. Vet. Res. 1980;41:219–223. - PubMed
- Duarte M., Laude H. Sequence of the spike protein of porcine epidemic diarrhea virus. J. Gen. Virol. 1994;75:1195–1200. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources