Using molecular classification to predict gains in maximal aerobic capacity following endurance exercise training in humans - PubMed (original) (raw)
. 2010 Jun;108(6):1487-96.
doi: 10.1152/japplphysiol.01295.2009. Epub 2010 Feb 4.
Steen Knudsen, Tuomo Rankinen, Lauren G Koch, Mark Sarzynski, Thomas Jensen, Pernille Keller, Camilla Scheele, Niels B J Vollaard, Søren Nielsen, Thorbjörn Akerström, Ormond A MacDougald, Eva Jansson, Paul L Greenhaff, Mark A Tarnopolsky, Luc J C van Loon, Bente K Pedersen, Carl Johan Sundberg, Claes Wahlestedt, Steven L Britton, Claude Bouchard
Affiliations
- PMID: 20133430
- PMCID: PMC2886694
- DOI: 10.1152/japplphysiol.01295.2009
Using molecular classification to predict gains in maximal aerobic capacity following endurance exercise training in humans
James A Timmons et al. J Appl Physiol (1985). 2010 Jun.
Abstract
A low maximal oxygen consumption (VO2max) is a strong risk factor for premature mortality. Supervised endurance exercise training increases VO2max with a very wide range of effectiveness in humans. Discovering the DNA variants that contribute to this heterogeneity typically requires substantial sample sizes. In the present study, we first use RNA expression profiling to produce a molecular classifier that predicts VO2max training response. We then hypothesized that the classifier genes would harbor DNA variants that contributed to the heterogeneous VO2max response. Two independent preintervention RNA expression data sets were generated (n=41 gene chips) from subjects that underwent supervised endurance training: one identified and the second blindly validated an RNA expression signature that predicted change in VO2max ("predictor" genes). The HERITAGE Family Study (n=473) was used for genotyping. We discovered a 29-RNA signature that predicted VO2max training response on a continuous scale; these genes contained approximately 6 new single-nucleotide polymorphisms associated with gains in VO2max in the HERITAGE Family Study. Three of four novel candidate genes from the HERITAGE Family Study were confirmed as RNA predictor genes (i.e., "reciprocal" RNA validation of a quantitative trait locus genotype), enhancing the performance of the 29-RNA-based predictor. Notably, RNA abundance for the predictor genes was unchanged by exercise training, supporting the idea that expression was preset by genetic variation. Regression analysis yielded a model where 11 single-nucleotide polymorphisms explained 23% of the variance in gains in VO2max, corresponding to approximately 50% of the estimated genetic variance for VO2max. In conclusion, combining RNA profiling with single-gene DNA marker association analysis yields a strongly validated molecular predictor with meaningful explanatory power. VO2max responses to endurance training can be predicted by measuring a approximately 30-gene RNA expression signature in muscle prior to training. The general approach taken could accelerate the discovery of genetic biomarkers, sufficiently discrete for diagnostic purposes, for a range of physiological and pharmacological phenotypes in humans.
Figures
Fig. 1.
An overview of the basic research strategy, along with the approximate sample sizes, used in this study. SNP, single-nucleotide polymorphism.
Fig. 2.
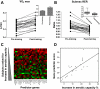
A and B: aerobic capacity and substrate oxidation characteristics of young sedentary human subjects before and after 6 wk of aerobic exercise training. Maximum oxygen consumption (V̇
o
2max) increased 14% (P < 0.0001) and submaximal exercise respiratory exchange ratio (RER) decreased, indicating greater reliance on lipid oxidation in vivo (P < 0.0001). Insets: geometric means ± SE pre- and posttraining. C and D: molecular predictor for aerobic adaptation to exercise training in humans involving 29 Affymetrix U133 Plus 2.0 probe sets. Logit-transformed normalized probe set intensities are correlated with ml/min increase in aerobic capacity (V̇
o
2max). C: green indicates higher gene expression, while red indicates lower gene expression. D: linear relationship between the sum of the expression of the molecular predictor genes and changes in aerobic capacity with endurance training derived from young sedentary subjects following 6 wk of constant-load aerobic exercise training (n = 24, _r_2 = 0.58, P < 0.00001).
Fig. 3.
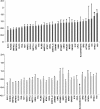
Skeletal muscle expression of all the predictor genes (based on RNA), which together explain ∼58% of the variance in exercise training-induced changes in V̇
o
2max in young sedentary human subjects (n = 24) before and after 6 wk of aerobic exercise training. Top: high responders following 6 wk of aerobic training; bottom: low responders following 6 wk of aerobic training. Expression values are generated using Affymetrix U133 Plus 2.0 gene chips (>47,000 transcripts), normalized using MAS5.0, and corrected for multiple comparisons using the significance of microarrays analysis methodology. This analysis strategy avoids issues with unstable housekeeping genes. Only H19, KLF4, OCT3, SMTNL2, and BTNL9 demonstrated a modest change in expression below a false discovery rate of 5% (∗), and not consistently in high and low responders. All other genes (>90% of predictor genes) were unchanged.
Fig. 4.
Genes identified as being able to predict which subjects demonstrate high response to exercise training formed a gene network found to be related to development. A: predictor genes (gray symbols with blue labeling) are not regulated by exercise (Table 1). Additional genes within the network represent genes known to interact with the predictor genes. B: genes shown in red are those that are increased by exercise training in group 1 (while those shown in blue are decreased). This demonstrates that most of the predictor genes, and many of those with which they interact in this analysis, are not responsive to regular exercise. Several members of the network are, however, known to be regulated by exercise at the protein level.
Comment in
- Does your (genetic) alphabet soup spell "runner"?
Bamman MM. Bamman MM. J Appl Physiol (1985). 2010 Jun;108(6):1452-3. doi: 10.1152/japplphysiol.00268.2010. Epub 2010 Mar 18. J Appl Physiol (1985). 2010. PMID: 20299619 No abstract available.
Similar articles
- Does your (genetic) alphabet soup spell "runner"?
Bamman MM. Bamman MM. J Appl Physiol (1985). 2010 Jun;108(6):1452-3. doi: 10.1152/japplphysiol.00268.2010. Epub 2010 Mar 18. J Appl Physiol (1985). 2010. PMID: 20299619 No abstract available. - Genomic predictors of the maximal O₂ uptake response to standardized exercise training programs.
Bouchard C, Sarzynski MA, Rice TK, Kraus WE, Church TS, Sung YJ, Rao DC, Rankinen T. Bouchard C, et al. J Appl Physiol (1985). 2011 May;110(5):1160-70. doi: 10.1152/japplphysiol.00973.2010. Epub 2010 Dec 23. J Appl Physiol (1985). 2011. PMID: 21183627 Free PMC article. - A transcriptional map of the impact of endurance exercise training on skeletal muscle phenotype.
Keller P, Vollaard NB, Gustafsson T, Gallagher IJ, Sundberg CJ, Rankinen T, Britton SL, Bouchard C, Koch LG, Timmons JA. Keller P, et al. J Appl Physiol (1985). 2011 Jan;110(1):46-59. doi: 10.1152/japplphysiol.00634.2010. Epub 2010 Oct 7. J Appl Physiol (1985). 2011. PMID: 20930125 Free PMC article. - The relationship between aerobic fitness and recovery from high intensity intermittent exercise.
Tomlin DL, Wenger HA. Tomlin DL, et al. Sports Med. 2001;31(1):1-11. doi: 10.2165/00007256-200131010-00001. Sports Med. 2001. PMID: 11219498 Review.
Cited by
- The magnitude of Yo-Yo test improvements following an aerobic training intervention are associated with total genotype score.
Pickering C, Kiely J, Suraci B, Collins D. Pickering C, et al. PLoS One. 2018 Nov 28;13(11):e0207597. doi: 10.1371/journal.pone.0207597. eCollection 2018. PLoS One. 2018. PMID: 30485313 Free PMC article. - Association Between Changes in Serum and Skeletal Muscle Metabolomics Profile With Maximum Power Output Gains in Response to Different Aerobic Training Programs: The Times Study.
Castro A, Duft RG, de Oliveira-Nunes SG, de Andrade ALL, Cavaglieri CR, Chacon-Mikahil MPT. Castro A, et al. Front Physiol. 2021 Oct 20;12:756618. doi: 10.3389/fphys.2021.756618. eCollection 2021. Front Physiol. 2021. PMID: 34744794 Free PMC article. - Physical Exercise as Personalized Medicine for Dementia Prevention?
Müllers P, Taubert M, Müller NG. Müllers P, et al. Front Physiol. 2019 May 29;10:672. doi: 10.3389/fphys.2019.00672. eCollection 2019. Front Physiol. 2019. PMID: 31244669 Free PMC article. - MicroRNAs in skeletal muscle and their regulation with exercise, ageing, and disease.
Zacharewicz E, Lamon S, Russell AP. Zacharewicz E, et al. Front Physiol. 2013 Sep 30;4:266. doi: 10.3389/fphys.2013.00266. Front Physiol. 2013. PMID: 24137130 Free PMC article. Review. - Effect of 8-week of dietary micronutrient supplementation on gene expression in elite handball athletes.
Molina-López J, Ricalde MAQ, Hernández BV, Planells A, Otero R, Planells E. Molina-López J, et al. PLoS One. 2020 May 1;15(5):e0232237. doi: 10.1371/journal.pone.0232237. eCollection 2020. PLoS One. 2020. PMID: 32357196 Free PMC article. Clinical Trial.
References
- Akimoto T, Ribar TJ, Williams RS, Yan Z. Skeletal muscle adaptation in response to voluntary running in Ca2+/calmodulin-dependent protein kinase IV-deficient mice. Am J Physiol Cell Physiol 287: C1311–C1319, 2004 - PubMed
- Baar K, Wende AR, Jones TE, Marison M, Nolte LA, Chen M, Kelly DP, Holloszy JO. Adaptations of skeletal muscle to exercise: rapid increase in the transcriptional coactivator PGC-1. FASEB J 16: 1879–1886, 2002 - PubMed
- Baldwin JE, Krebs H. The evolution of metabolic cycles. Nature 291: 381–382, 1981 - PubMed
- Blair SN, Kampert JB, Kohl HW, 3rd, Barlow CE, Macera CA, Paffenbarger RS, Jr, Gibbons LW. Influences of cardiorespiratory fitness and other precursors on cardiovascular disease and all-cause mortality in men and women. JAMA 276: 205–210, 1996 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- RR-17718/RR/NCRR NIH HHS/United States
- HL-45670/HL/NHLBI NIH HHS/United States
- HL-47321/HL/NHLBI NIH HHS/United States
- HL-47317/HL/NHLBI NIH HHS/United States
- HL-47327/HL/NHLBI NIH HHS/United States
- HL-47323/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases