Argonautes ALG-3 and ALG-4 are required for spermatogenesis-specific 26G-RNAs and thermotolerant sperm in Caenorhabditis elegans - PubMed (original) (raw)
Argonautes ALG-3 and ALG-4 are required for spermatogenesis-specific 26G-RNAs and thermotolerant sperm in Caenorhabditis elegans
Colin C Conine et al. Proc Natl Acad Sci U S A. 2010.
Abstract
Gametogenesis is a thermosensitive process in numerous metazoans, ranging from worms to man. In Caenorhabditis elegans, a variety of RNA-binding proteins that associate with germ-line nuage (P granules), including the Piwi-clade argonaute PRG-1, have been implicated in maintaining fertility at elevated temperature. Here we describe the role of two AGO-class paralogs, alg-3 (T22B3.2) and alg-4 (ZK757.3), in promoting thermotolerant male fertility. A rescuing GFP::alg-3 transgene is localized to P granules beginning at the late pachytene stage of male gametogenesis. alg-3/4 double mutants lack a subgroup of small RNAs, the 26G-RNAs which target and appear to down-regulate numerous spermatogenesis-expressed mRNAs. These findings add to a growing number of AGO pathways required for thermotolerant fertility in C. elegans and support a model in which AGOs and their small RNA cofactors function to promote robustness in gene-expression networks.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
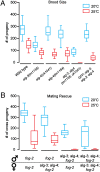
alg-3/4 mutants exhibit temperature-sensitive sterility associated with the male germline. (A and B) Box-and-whiskers plots of brood size in wild-type and mutant strains as indicated for (n > 20) animals cultured at 20 °C (blue) and 25 °C (red). In these and all subsequent box-and-whisker plots, the top and bottom ends of each box represent the 75th and 25th percentile, respectively; the line in the middle represents the median value; and the extended lines illustrate the range (highest and lowest value). The temperature of cultivation is indicated by color. (A) Self-crosses; (B) crosses between fog-2 (wild-type) and alg-3/4;fog-2 (mutant) worms.
Fig. 2.
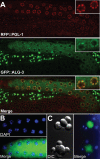
GFP::ALG-3 is expressed during spermatogenesis. (A–C) Micrographs of ALG-3::GFP (green), nuclear staining/DAPI (blue), and PGL-1::RFP (red). (A) Confocal images in a young adult hermaphrodite. (B) Onset of expression in the proximal region of a male gonad. (C) Nomarski and fluorescence images of spermatids attached to residual bodies.
Fig. 3.
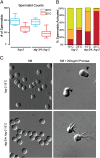
alg-3/4 mutants exhibit defects in sperm activation. (A) Box-and-whisker plots of spermatid counts performed on (n > 20) wild-type (fog-2) and alg-3/4;fog-2 animals (as indicated) cultured at 20 °C (blue) or 25 °C (red). (B) Graphic depiction of spermatid activation, illustrating the percent of spermatids with pseudopods (green), spikes (orange), or unactivated (red) (n > 200). (C) Nomarski images of wild-type (fog-2) and alg-3/4;fog-2 spermatids before (Left) and after (Right) activation. Black arrowheads denote pseudopods.
Fig. 4.
Analysis of 26G-RNA expression and targeting. (A) Length and first-nucleotide distribution of deep-sequencing reads from wild-type (fog-2) (Left), and alg-3/4; fog-2 (Right). (B) Two-point plots comparing the relative proportions of various small RNA classes (as indicated by color) in wild-type (fog-2) (Left) and alg-3/4; fog-2 mutants (Right) for 26nt long reads. (C) Box-and-whisker plots depicting relative mRNA levels in microarray assays on alg-3/4 (mutant) and N2 (wild-type) populations. Here and in Fig. 5, the y axis represents the relative mRNA levels [measured as (alg-3/4 mutant value divided by [wild-type plus alg-3/4 mutant values]) for any given locus]. Dotted lines indicate the values corresponding to 2-fold enrichment (a value of 0.66) or depletion (a value of 0.33). (D) RT-qPCR measurement of target mRNAs up-regulated (bold type) or not regulated (regular type) based on microarray analysis. k02e2.6 is an _ergo-1_-dependent 26G-RNA target. (E) Schematic representation of 26G-RNA targets defined using using a cutoff of 10 reads per million. (F) Northern blot analysis of small RNAs in wild-type and various mutant backgrounds as indicated. SL1 and mir-66 are loading controls.
Fig. 5.
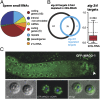
Analysis of Small RNA pathways in mature sperm. (A) Pie chart representing the distribution of different classes of small RNA present in isolated spermatids. The 26nt RNAs represent less than 2% of the total small RNA reads. (B) alg-3/4 targets are also 22G-RNA targets. The Venn diagram shows intersection between regulated alg-3/4 targets (based on microarray) and targets that are depleted 2-fold or greater of 22G-RNAs. The box-and-whisker diagram shows depletion of 22G-RNAs relative to wild-type on alg-3/4 targets. y axis represents the relative proportion of reads (measure as alg-3/4 mutant value divided by wild type value plus alg-3/4 mutant value). (C) GFP::WAGO-1 expression in an adult male (upper panel). Spermatids are marked with a white arrow head. GFP::WAGO-1 expression in individual spermatids, also stained with DAPI (blue, lower panel).
Fig. 6.
Model of ALG-3/4 and WAGO-1 expression during sperm development. (A) Depiction of spermatogenesis and spermiogenesis, with ALG-3/4 and WAGO-1 expression indicated by the red and green bars (respectively), as well as a model for the biogenesis of 26G and 22G RNAs. The schematic in B illustrates a potential role for ALG-3/4 in lowering target-mRNA levels to increase robustness to temperature.
Similar articles
- Biogenesis of C. elegans spermatogenesis small RNAs is initiated by a zc3h12a-like ribonuclease.
Tsai HY, Cheng HT, Tsai YT. Tsai HY, et al. Sci Adv. 2022 Aug 12;8(32):eabm0699. doi: 10.1126/sciadv.abm0699. Epub 2022 Aug 10. Sci Adv. 2022. PMID: 35947655 Free PMC article. - ALG-5 is a miRNA-associated Argonaute required for proper developmental timing in the Caenorhabditis elegans germline.
Brown KC, Svendsen JM, Tucci RM, Montgomery BE, Montgomery TA. Brown KC, et al. Nucleic Acids Res. 2017 Sep 6;45(15):9093-9107. doi: 10.1093/nar/gkx536. Nucleic Acids Res. 2017. PMID: 28645154 Free PMC article. - Small RNA-mediated genetic switches coordinate ALG-3/4 small RNA pathway function.
Sen T, McCormick C, Rogers AK. Sen T, et al. Nucleic Acids Res. 2024 Sep 9;52(16):9431-9449. doi: 10.1093/nar/gkae586. Nucleic Acids Res. 2024. PMID: 38967024 Free PMC article. - [Recent advances in the study of spermatogenesis and fertilization in Caenorhabditis elegans].
Wang B. Wang B. Yi Chuan. 2008 Jun;30(6):677-86. doi: 10.3724/sp.j.1005.2008.00677. Yi Chuan. 2008. PMID: 18550488 Review. Chinese. - Spermatogenesis.
L'Hernault SW. L'Hernault SW. WormBook. 2006 Feb 20:1-14. doi: 10.1895/wormbook.1.85.1. WormBook. 2006. PMID: 18050478 Free PMC article. Review.
Cited by
- A conserved upstream motif orchestrates autonomous, germline-enriched expression of Caenorhabditis elegans piRNAs.
Billi AC, Freeberg MA, Day AM, Chun SY, Khivansara V, Kim JK. Billi AC, et al. PLoS Genet. 2013;9(3):e1003392. doi: 10.1371/journal.pgen.1003392. Epub 2013 Mar 14. PLoS Genet. 2013. PMID: 23516384 Free PMC article. - MUT-16 promotes formation of perinuclear mutator foci required for RNA silencing in the C. elegans germline.
Phillips CM, Montgomery TA, Breen PC, Ruvkun G. Phillips CM, et al. Genes Dev. 2012 Jul 1;26(13):1433-44. doi: 10.1101/gad.193904.112. Epub 2012 Jun 19. Genes Dev. 2012. PMID: 22713602 Free PMC article. - RNAi pathways contribute to developmental history-dependent phenotypic plasticity in C. elegans.
Hall SE, Chirn GW, Lau NC, Sengupta P. Hall SE, et al. RNA. 2013 Mar;19(3):306-19. doi: 10.1261/rna.036418.112. Epub 2013 Jan 17. RNA. 2013. PMID: 23329696 Free PMC article. - Plastic germline reprogramming of heritable small RNAs enables maintenance or erasure of epigenetic memories.
Houri-Ze'evi L, Rechavi O. Houri-Ze'evi L, et al. RNA Biol. 2016 Dec;13(12):1212-1217. doi: 10.1080/15476286.2016.1229732. Epub 2016 Sep 3. RNA Biol. 2016. PMID: 27592591 Free PMC article. - Spatiotemporal Gene Expression Analysis of the Caenorhabditis elegans Germline Uncovers a Syncytial Expression Switch.
Tzur YB, Winter E, Gao J, Hashimshony T, Yanai I, Colaiácovo MP. Tzur YB, et al. Genetics. 2018 Oct;210(2):587-605. doi: 10.1534/genetics.118.301315. Epub 2018 Aug 9. Genetics. 2018. PMID: 30093412 Free PMC article.
References
- Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9:22–32. - PubMed
- Tolia NH, Joshua-Tor L. Slicer and the argonautes. Nat Chem Biol. 2007;3:36–43. - PubMed
- Yigit E, et al. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell. 2006;127:747–757. - PubMed
- Grishok A, et al. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials