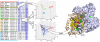Protein interactions and ligand binding: from protein subfamilies to functional specificity - PubMed (original) (raw)
Protein interactions and ligand binding: from protein subfamilies to functional specificity
Antonio Rausell et al. Proc Natl Acad Sci U S A. 2010.
Abstract
The divergence accumulated during the evolution of protein families translates into their internal organization as subfamilies, and it is directly reflected in the characteristic patterns of differentially conserved residues. These specifically conserved positions in protein subfamilies are known as "specificity determining positions" (SDPs). Previous studies have limited their analysis to the study of the relationship between these positions and ligand-binding specificity, demonstrating significant yet limited predictive capacity. We have systematically extended this observation to include the role of differential protein interactions in the segregation of protein subfamilies and explored in detail the structural distribution of SDPs at protein interfaces. Our results show the extensive influence of protein interactions in the evolution of protein families and the widespread association of SDPs with protein interfaces. The combined analysis of SDPs in interfaces and ligand-binding sites provides a more complete picture of the organization of protein families, constituting the necessary framework for a large scale analysis of the evolution of protein function.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
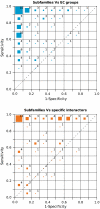
Fig. 1.
Correspondence between the different subfamilies, and the EC groups (Top) and specific interactors (Bottom) for each protein family represented in the ROC space, where the distribution of the families is shown as a bidimensional histogram. The size of the colored boxes in each bin of the ROC space represents the percentage of protein families they contain, whereas the number shows the actual percentage. For the sake of simplicity, percentage values are rounded to the nearest integer (so that they may not add up to 100).
Fig. 2.
Workflow implemented to simultaneously detect the protein subfamilies and those residues responsible for such segregation (SDPs). This process is depicted for the class III aminotransferase family (Pfam PF00202). The homodimeric structure of the human ornithine aminotransferase (PDB 1oat) bound to Pyridoxal-5’-phosphate (in red spheres) is represented. The two subunits of the complex are shown as brown cartoon representation and with a gray surface. SDPs are highlighted in a yellow/violet spacefill and with a green surface. The figure was generated with Pymol (pymol.sourceforge.com).
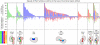
Fig. 3.
Percentage of SDPs in the functional regions (Top) compared to the corresponding percentage of protein residues in these functional regions (medium) in each Pfam family. The data are grouped according to the type of functional region detected in each family (Bottom), whereas the number of families in each category is shown in parentheses. Ligand-binding sites shown in blue, heterodimeric interfaces in green, homodimeric interface in red, and their combinations in yellow. The intrachain interfaces have been omitted for the sake of simplicity.
Similar articles
- Evolution of protein interactions: from interactomes to interfaces.
Andreani J, Guerois R. Andreani J, et al. Arch Biochem Biophys. 2014 Jul 15;554:65-75. doi: 10.1016/j.abb.2014.05.010. Epub 2014 May 20. Arch Biochem Biophys. 2014. PMID: 24853495 Review. - Joint evolutionary trees: a large-scale method to predict protein interfaces based on sequence sampling.
Engelen S, Trojan LA, Sacquin-Mora S, Lavery R, Carbone A. Engelen S, et al. PLoS Comput Biol. 2009 Jan;5(1):e1000267. doi: 10.1371/journal.pcbi.1000267. Epub 2009 Jan 23. PLoS Comput Biol. 2009. PMID: 19165315 Free PMC article. - An evolutionary trace method defines binding surfaces common to protein families.
Lichtarge O, Bourne HR, Cohen FE. Lichtarge O, et al. J Mol Biol. 1996 Mar 29;257(2):342-58. doi: 10.1006/jmbi.1996.0167. J Mol Biol. 1996. PMID: 8609628 - Combining specificity determining and conserved residues improves functional site prediction.
Kalinina OV, Gelfand MS, Russell RB. Kalinina OV, et al. BMC Bioinformatics. 2009 Jun 9;10:174. doi: 10.1186/1471-2105-10-174. BMC Bioinformatics. 2009. PMID: 19508719 Free PMC article. - Structure function relations in PDZ-domain-containing proteins: Implications for protein networks in cellular signalling.
Manjunath GP, Ramanujam PL, Galande S. Manjunath GP, et al. J Biosci. 2018 Mar;43(1):155-171. J Biosci. 2018. PMID: 29485124 Review.
Cited by
- Novel Computational Protocols for Functionally Classifying and Characterising Serine Beta-Lactamases.
Lee D, Das S, Dawson NL, Dobrijevic D, Ward J, Orengo C. Lee D, et al. PLoS Comput Biol. 2016 Jun 22;12(6):e1004926. doi: 10.1371/journal.pcbi.1004926. eCollection 2016 Jun. PLoS Comput Biol. 2016. PMID: 27332861 Free PMC article. - Networks of high mutual information define the structural proximity of catalytic sites: implications for catalytic residue identification.
Marino Buslje C, Teppa E, Di Doménico T, Delfino JM, Nielsen M. Marino Buslje C, et al. PLoS Comput Biol. 2010 Nov 4;6(11):e1000978. doi: 10.1371/journal.pcbi.1000978. PLoS Comput Biol. 2010. PMID: 21079665 Free PMC article. - The functional importance of co-evolving residues in proteins.
Sandler I, Zigdon N, Levy E, Aharoni A. Sandler I, et al. Cell Mol Life Sci. 2014 Feb;71(4):673-82. doi: 10.1007/s00018-013-1458-2. Epub 2013 Sep 1. Cell Mol Life Sci. 2014. PMID: 23995987 Free PMC article. Review. - Conserved differences in protein sequence determine the human pathogenicity of Ebolaviruses.
Pappalardo M, Juliá M, Howard MJ, Rossman JS, Michaelis M, Wass MN. Pappalardo M, et al. Sci Rep. 2016 Mar 24;6:23743. doi: 10.1038/srep23743. Sci Rep. 2016. PMID: 27009368 Free PMC article. - Local energetic frustration conservation in protein families and superfamilies.
Freiberger MI, Ruiz-Serra V, Pontes C, Romero-Durana M, Galaz-Davison P, Ramírez-Sarmiento CA, Schuster CD, Marti MA, Wolynes PG, Ferreiro DU, Parra RG, Valencia A. Freiberger MI, et al. Nat Commun. 2023 Dec 16;14(1):8379. doi: 10.1038/s41467-023-43801-2. Nat Commun. 2023. PMID: 38104123 Free PMC article.
References
- Orengo CA, Thornton JM. Protein families and their evolution-a structural perspective. Annu Rev Biochem. 2005;74:867–900. - PubMed
- Koonin EV, Wolf YI, Karev GP. The structure of the protein universe and genome evolution. Nature. 2002;420:218–223. - PubMed
- Zuckerkandl E, Pauling L. Molecules as documents of evolutionary history. J Theor Biol. 1965;8:357–366. - PubMed
- Valdar WS. Scoring residue conservation. Proteins. 2002;48(2):227–241. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources