Latent herpesvirus infection arms NK cells - PubMed (original) (raw)
Latent herpesvirus infection arms NK cells
Douglas W White et al. Blood. 2010.
Abstract
Natural killer (NK) cells were identified by their ability to kill target cells without previous sensitization. However, without an antecedent "arming" event, NK cells can recognize, but are not equipped to kill, target cells. How NK cells become armed in vivo in healthy hosts is unclear. Because latent herpesviruses are highly prevalent and alter multiple aspects of host immunity, we hypothesized that latent herpesvirus infection would arm NK cells. Here we show that NK cells from mice latently infected with Murid herpesvirus 4 (MuHV-4) were armed as evidenced by increased granzyme B protein expression, cytotoxicity, and interferon-gamma production. NK-cell arming occurred rapidly in the latently infected host and did not require acute viral infection. Furthermore, NK cells armed by latent infection protected the host against a lethal lymphoma challenge. Thus, the immune environment created by latent herpesvirus infection provides a mechanism whereby host NK-cell function is enhanced in vivo.
Figures
Figure 1
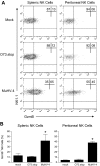
Latent infection with MuHV-4 arms NK cells with GzmB protein. Mice were inoculated with media (mock), MuHV-4, or latency-defective MuHV-4 (O73.stop) 31 days before flow cytometric analysis of GzmB expression in NK1.1+CD3− NK cells from the spleen and peritoneal cavity. (A) Representative density plots demonstrating GzmB expression within NK1.1+CD3− NK cells. (B) Pooled data (10 mice per condition) showing the mean ± SD percentage of NK cells that express GzmB protein. These data are representative of 4 independent experiments, *P < .009 (MuHV-4 vs O73.stop or mock).
Figure 2
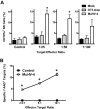
Latent infection with MuHV-4 enhances NK-cell degranulation and cytotoxicity. (A) Latent infection with MuHV-4 increases NK-cell degranulation. Peritoneal cells from mock, O73.stop, or MuHV-4 latently infected mice were cultured ex vivo with YAC-1 target cells at the indicated target:effector ratios for 2 hours, followed by flow cytometric analysis of CD107a expression on the surface of NK1.1+CD3− NK cells. Control mice were incubated without targets. Bars represent the mean ± SD of 5 to 10 mice per group. *P < .032 (MuHV-4 compared with mock or O73.stop). These data are representative of 2 independent experiments with 15 to 20 total mice per condition. (B) Latent infection with MuHV-4 induces cytotoxic activity by NK cells. Peritoneal cells from control (naive) mice and mice infected 28 days previously with MuHV-4 were enriched for NK cells and then coincubated with RMA-S target cells for 4 hours at the indicated effector:target ratios before analysis of target cell death by 7-AAD. *P < .003 (MuHV-4 vs control). These data are representative of 2 independent experiments with 20 to 30 mice per group per experiment.
Figure 3
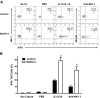
NK cells from mice latently infected with MuHV-4 have increased capacity to produce IFN-γ protein. Peritoneal cells from latently infected or control (naive or mock-infected) mice were stained immediately after harvest (No Cx) or after culture ex vivo with phosphate-buffered saline (PBS), IL-12 plus IL-15, or plate-bound antibodies against NK1.1 (Anti-NK1.1). (A) Representative flow cytometric density plots showing IFN-γ expression in NK cells. (B) Pooled data from 10 mice per group showing the mean ± SD percentage of NK cells that express IFN-γ. These data are representative of 3 independent experiments with 10 mice per group per experiment. *P <.001 (MuHV-4 vs control).
Figure 4
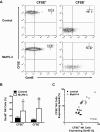
NK cells express increased GzmB protein after short-term exposure to the latent MuHV-4 environment in vivo. Latently infected mice (28 days after infection with MuHV-4) were injected intraperitoneally with CFSE-labeled splenocytes from B6.RAG1−/− donors as a source of naive NK cells. Mock-infected and naive recipient mice were used as controls. At 72 hours peritoneal cells were harvested and analyzed for CFSE and GzmB expression in NK1.1+CD3− NK cells. (A) Representative flow cytometric density plots illustrating the expression of GzmB protein in transferred (CFSE+) versus endogenous (CFSE−) NK cells. (B) Pooled data from 2 independent experiments showing the mean ± SD percentage of NK cells that express GzmB. *P = .027 and **P < .001 (MuHV-4 vs control, 11-14 mice per group). (C) Correlation of GzmB protein expression between transferred versus endogenous NK cells in each mouse. Each dot represents one mouse. Correlation coefficient (r2) = 0.9 in latently infected hosts.
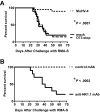
Figure 5
NK cells in latently infected mice protect against a lethal RMA-S lymphoma challenge. (A) Mice were inoculated with medium (mock), MuHV-4, or latency-defective MuHV-4 (O73.stop) 29 days before injection with 103 RMA-S cells intraperitoneally and then followed for survival. These data are pooled from 3 independent experiments with a total of 20 mice per group. *P < .001 (MuHV-4 vs either control). (B) Mice were inoculated with MuHV-4 35 days before injection with anti-NK1.1 or control monoclonal antibody (mAb). One day later, all animals were challenged with RMA-S as in panel A. Antibody injections were continued every 6 to 8 days for the duration of the experiment. These data are pooled from 2 independent experiments with a total of 10 mice per group. *P < .001 (anti-NK1.1 vs control).
Comment in
- Latent herpesviruses: aligning human and murine NK cells.
Orange JS. Orange JS. Blood. 2010 Jun 3;115(22):4321-2. doi: 10.1182/blood-2010-03-271361. Blood. 2010. PMID: 20522715 No abstract available.
Similar articles
- Type I Interferons and NK Cells Restrict Gammaherpesvirus Lymph Node Infection.
Lawler C, Tan CS, Simas JP, Stevenson PG. Lawler C, et al. J Virol. 2016 Sep 29;90(20):9046-57. doi: 10.1128/JVI.01108-16. Print 2016 Oct 15. J Virol. 2016. PMID: 27466430 Free PMC article. - Indirect CD4+ T cell protection against mouse gamma-herpesvirus infection via interferon gamma.
Xie W, Bruce K, Belz GT, Farrell HE, Stevenson PG. Xie W, et al. J Virol. 2024 May 14;98(5):e0049324. doi: 10.1128/jvi.00493-24. Epub 2024 Apr 5. J Virol. 2024. PMID: 38578092 Free PMC article. - A CD4+ T Cell-NK Cell Axis of Gammaherpesvirus Control.
Lawler C, Stevenson PG. Lawler C, et al. J Virol. 2020 Jan 17;94(3):e01545-19. doi: 10.1128/JVI.01545-19. Print 2020 Jan 17. J Virol. 2020. PMID: 31694958 Free PMC article. - Immune control of mammalian gamma-herpesviruses: lessons from murid herpesvirus-4.
Stevenson PG, Simas JP, Efstathiou S. Stevenson PG, et al. J Gen Virol. 2009 Oct;90(Pt 10):2317-2330. doi: 10.1099/vir.0.013300-0. Epub 2009 Jul 15. J Gen Virol. 2009. PMID: 19605591 Review. - Herpesvirus Evasion of Natural Killer Cells.
De Pelsmaeker S, Romero N, Vitale M, Favoreel HW. De Pelsmaeker S, et al. J Virol. 2018 May 14;92(11):e02105-17. doi: 10.1128/JVI.02105-17. Print 2018 Jun 1. J Virol. 2018. PMID: 29540598 Free PMC article. Review.
Cited by
- Metagenomic analysis of intestinal mucosa revealed a specific eukaryotic gut virome signature in early-diagnosed inflammatory bowel disease.
Ungaro F, Massimino L, Furfaro F, Rimoldi V, Peyrin-Biroulet L, D'Alessio S, Danese S. Ungaro F, et al. Gut Microbes. 2019;10(2):149-158. doi: 10.1080/19490976.2018.1511664. Epub 2018 Sep 25. Gut Microbes. 2019. PMID: 30252582 Free PMC article. - Preactivation with IL-12, IL-15, and IL-18 induces CD25 and a functional high-affinity IL-2 receptor on human cytokine-induced memory-like natural killer cells.
Leong JW, Chase JM, Romee R, Schneider SE, Sullivan RP, Cooper MA, Fehniger TA. Leong JW, et al. Biol Blood Marrow Transplant. 2014 Apr;20(4):463-73. doi: 10.1016/j.bbmt.2014.01.006. Epub 2014 Jan 13. Biol Blood Marrow Transplant. 2014. PMID: 24434782 Free PMC article. - Herpesviruses: Harmonious Pathogens but Relevant Cofactors in Other Diseases?
Sehrawat S, Kumar D, Rouse BT. Sehrawat S, et al. Front Cell Infect Microbiol. 2018 May 25;8:177. doi: 10.3389/fcimb.2018.00177. eCollection 2018. Front Cell Infect Microbiol. 2018. PMID: 29888215 Free PMC article. Review. - Latent cytomegalovirus infection enhances anti-tumour cytotoxicity through accumulation of NKG2C+ NK cells in healthy humans.
Bigley AB, Rezvani K, Shah N, Sekine T, Balneger N, Pistillo M, Agha N, Kunz H, O'Connor DP, Bollard CM, Simpson RJ. Bigley AB, et al. Clin Exp Immunol. 2016 Aug;185(2):239-51. doi: 10.1111/cei.12785. Clin Exp Immunol. 2016. PMID: 26940026 Free PMC article. - Primary Immunodeficiency and Cancer Predisposition Revisited: Embedding Two Closely Related Concepts Into an Integrative Conceptual Framework.
Haas OA. Haas OA. Front Immunol. 2019 Feb 12;9:3136. doi: 10.3389/fimmu.2018.03136. eCollection 2018. Front Immunol. 2019. PMID: 30809233 Free PMC article. Review.
References
- Herberman RB, Nunn ME, Lavrin DH. Natural cytotoxic reactivity of mouse lymphoid cells against syngeneic acid allogeneic tumors. I. Distribution of reactivity and specificity. Int J Cancer. 1975;16(2):216–229. - PubMed
- Kiessling R, Klein E, Wigzell H. “Natural” killer cells in the mouse. I. Cytotoxic cells with specificity for mouse Moloney leukemia cells. Specificity and distribution according to genotype. Eur J Immunol. 1975;5(2):112–117. - PubMed
- Biron CA, Brossay L. NK cells and NKT cells in innate defense against viral infections. Curr Opin Immunol. 2001;13(4):458–464. - PubMed
- Cerwenka A, Lanier LL. Natural killer cells, viruses and cancer. Nat Rev Immunol. 2001;1(1):41–49. - PubMed
- Yokoyama WM, Kim S, French AR. The dynamic life of natural killer cells. Annu Rev Immunol. 2004;22:405–429. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 DK049786/DK/NIDDK NIH HHS/United States
- R01DK49786/DK/NIDDK NIH HHS/United States
- R01 CA074730/CA/NCI NIH HHS/United States
- K08HL093299/HL/NHLBI NIH HHS/United States
- K08 AI079011/AI/NIAID NIH HHS/United States
- U54 AI057160/AI/NIAID NIH HHS/United States
- R01CA096511/CA/NCI NIH HHS/United States
- R01 CA096511/CA/NCI NIH HHS/United States
- R01CA074730/CA/NCI NIH HHS/United States
- K08 HL093299/HL/NHLBI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- K08AI079011/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases